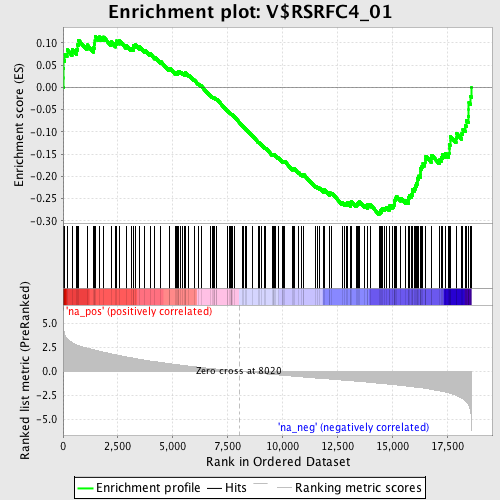
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$RSRFC4_01 |
| Enrichment Score (ES) | -0.2859427 |
| Normalized Enrichment Score (NES) | -1.451008 |
| Nominal p-value | 0.006369427 |
| FDR q-value | 0.97550935 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PIK3R3 | 12 | 4.890 | 0.0210 | No | ||
| 2 | PYY | 14 | 4.827 | 0.0424 | No | ||
| 3 | RG9MTD2 | 32 | 4.258 | 0.0604 | No | ||
| 4 | POU4F2 | 82 | 3.788 | 0.0745 | No | ||
| 5 | HOXB4 | 180 | 3.412 | 0.0844 | No | ||
| 6 | NFAT5 | 408 | 2.982 | 0.0853 | No | ||
| 7 | ANKMY2 | 623 | 2.757 | 0.0860 | No | ||
| 8 | WBP4 | 668 | 2.716 | 0.0956 | No | ||
| 9 | CUL3 | 707 | 2.691 | 0.1055 | No | ||
| 10 | TOB2 | 1091 | 2.424 | 0.0955 | No | ||
| 11 | TPP2 | 1392 | 2.244 | 0.0892 | No | ||
| 12 | RAP2C | 1416 | 2.232 | 0.0979 | No | ||
| 13 | MBNL2 | 1452 | 2.211 | 0.1058 | No | ||
| 14 | SLAMF1 | 1459 | 2.209 | 0.1152 | No | ||
| 15 | CUTL1 | 1662 | 2.094 | 0.1136 | No | ||
| 16 | CFL2 | 1833 | 2.009 | 0.1133 | No | ||
| 17 | STAG2 | 2183 | 1.825 | 0.1025 | No | ||
| 18 | CDC42EP3 | 2401 | 1.721 | 0.0983 | No | ||
| 19 | ATP1B2 | 2430 | 1.708 | 0.1044 | No | ||
| 20 | SLC26A6 | 2560 | 1.653 | 0.1047 | No | ||
| 21 | CASQ2 | 2869 | 1.511 | 0.0947 | No | ||
| 22 | GRIN2B | 3102 | 1.411 | 0.0884 | No | ||
| 23 | LUC7L | 3195 | 1.371 | 0.0895 | No | ||
| 24 | FGF12 | 3212 | 1.365 | 0.0947 | No | ||
| 25 | PLAGL2 | 3302 | 1.330 | 0.0958 | No | ||
| 26 | HAPLN1 | 3487 | 1.260 | 0.0914 | No | ||
| 27 | AMPD1 | 3731 | 1.163 | 0.0834 | No | ||
| 28 | MEOX2 | 3961 | 1.079 | 0.0758 | No | ||
| 29 | RASGRP3 | 4184 | 1.005 | 0.0682 | No | ||
| 30 | CNTN1 | 4458 | 0.919 | 0.0575 | No | ||
| 31 | EPHA7 | 4832 | 0.807 | 0.0408 | No | ||
| 32 | WFDC1 | 4870 | 0.798 | 0.0424 | No | ||
| 33 | ADCYAP1 | 5136 | 0.712 | 0.0311 | No | ||
| 34 | LCP1 | 5160 | 0.706 | 0.0330 | No | ||
| 35 | EYA1 | 5231 | 0.687 | 0.0323 | No | ||
| 36 | CENTD1 | 5239 | 0.685 | 0.0349 | No | ||
| 37 | LZTS2 | 5270 | 0.677 | 0.0363 | No | ||
| 38 | GPC4 | 5372 | 0.640 | 0.0337 | No | ||
| 39 | SIPA1L1 | 5450 | 0.620 | 0.0323 | No | ||
| 40 | ART5 | 5552 | 0.593 | 0.0294 | No | ||
| 41 | HIST1H2AA | 5560 | 0.592 | 0.0317 | No | ||
| 42 | SLC8A3 | 5566 | 0.589 | 0.0340 | No | ||
| 43 | ZHX2 | 5732 | 0.541 | 0.0275 | No | ||
| 44 | FOXP1 | 5973 | 0.483 | 0.0166 | No | ||
| 45 | STAC | 6176 | 0.429 | 0.0076 | No | ||
| 46 | KPNA3 | 6286 | 0.403 | 0.0034 | No | ||
| 47 | MGST3 | 6703 | 0.302 | -0.0178 | No | ||
| 48 | MYL3 | 6813 | 0.276 | -0.0225 | No | ||
| 49 | ASB16 | 6870 | 0.263 | -0.0243 | No | ||
| 50 | PANK1 | 6882 | 0.261 | -0.0238 | No | ||
| 51 | IRS1 | 6920 | 0.249 | -0.0247 | No | ||
| 52 | PAX3 | 6985 | 0.235 | -0.0271 | No | ||
| 53 | MUSK | 6990 | 0.234 | -0.0263 | No | ||
| 54 | WSB2 | 7509 | 0.115 | -0.0538 | No | ||
| 55 | FST | 7512 | 0.114 | -0.0534 | No | ||
| 56 | HOXD4 | 7586 | 0.100 | -0.0570 | No | ||
| 57 | COL8A1 | 7640 | 0.088 | -0.0594 | No | ||
| 58 | HRASLS | 7685 | 0.079 | -0.0615 | No | ||
| 59 | BZW2 | 7691 | 0.078 | -0.0614 | No | ||
| 60 | MYH3 | 7695 | 0.078 | -0.0612 | No | ||
| 61 | FBXW11 | 7737 | 0.066 | -0.0631 | No | ||
| 62 | LECT1 | 7824 | 0.049 | -0.0676 | No | ||
| 63 | CAV3 | 8168 | -0.032 | -0.0860 | No | ||
| 64 | CLDN14 | 8207 | -0.039 | -0.0879 | No | ||
| 65 | EDG1 | 8326 | -0.068 | -0.0940 | No | ||
| 66 | PDGFRA | 8362 | -0.075 | -0.0956 | No | ||
| 67 | THBS2 | 8649 | -0.134 | -0.1105 | No | ||
| 68 | TNNI3K | 8898 | -0.185 | -0.1231 | No | ||
| 69 | HOXC4 | 8957 | -0.197 | -0.1254 | No | ||
| 70 | ESR1 | 9054 | -0.220 | -0.1296 | No | ||
| 71 | ITSN1 | 9194 | -0.250 | -0.1360 | No | ||
| 72 | SSH3 | 9240 | -0.262 | -0.1373 | No | ||
| 73 | TRDN | 9551 | -0.329 | -0.1527 | No | ||
| 74 | TMEPAI | 9583 | -0.337 | -0.1529 | No | ||
| 75 | ADAM11 | 9605 | -0.341 | -0.1525 | No | ||
| 76 | ACTA1 | 9616 | -0.343 | -0.1515 | No | ||
| 77 | SH3GLB2 | 9684 | -0.356 | -0.1535 | No | ||
| 78 | TNNC1 | 9808 | -0.381 | -0.1585 | No | ||
| 79 | PCDH9 | 10005 | -0.414 | -0.1673 | No | ||
| 80 | MYL2 | 10016 | -0.416 | -0.1660 | No | ||
| 81 | MEF2C | 10048 | -0.422 | -0.1658 | No | ||
| 82 | STOML2 | 10111 | -0.434 | -0.1673 | No | ||
| 83 | SV2A | 10462 | -0.500 | -0.1840 | No | ||
| 84 | ELAVL4 | 10495 | -0.507 | -0.1835 | No | ||
| 85 | DNAJA4 | 10528 | -0.514 | -0.1829 | No | ||
| 86 | MRPS23 | 10735 | -0.552 | -0.1917 | No | ||
| 87 | DKK2 | 10858 | -0.574 | -0.1957 | No | ||
| 88 | TRPS1 | 10949 | -0.593 | -0.1980 | No | ||
| 89 | HAO1 | 10956 | -0.595 | -0.1957 | No | ||
| 90 | BAI3 | 11517 | -0.699 | -0.2229 | No | ||
| 91 | ATF3 | 11613 | -0.714 | -0.2249 | No | ||
| 92 | PRDM1 | 11699 | -0.726 | -0.2263 | No | ||
| 93 | KCNN1 | 11880 | -0.757 | -0.2327 | No | ||
| 94 | HDAC9 | 11899 | -0.760 | -0.2303 | No | ||
| 95 | GCK | 12135 | -0.803 | -0.2395 | No | ||
| 96 | S100A4 | 12142 | -0.805 | -0.2362 | No | ||
| 97 | GPR153 | 12240 | -0.824 | -0.2378 | No | ||
| 98 | TNNI2 | 12713 | -0.909 | -0.2594 | No | ||
| 99 | ZFPM2 | 12845 | -0.934 | -0.2623 | No | ||
| 100 | KBTBD5 | 12917 | -0.947 | -0.2620 | No | ||
| 101 | FBXO40 | 12950 | -0.955 | -0.2595 | No | ||
| 102 | NDRG2 | 13101 | -0.980 | -0.2633 | No | ||
| 103 | DIRAS1 | 13102 | -0.980 | -0.2589 | No | ||
| 104 | RALY | 13132 | -0.985 | -0.2561 | No | ||
| 105 | CYP26B1 | 13358 | -1.032 | -0.2637 | No | ||
| 106 | NEDD4 | 13400 | -1.039 | -0.2614 | No | ||
| 107 | CASQ1 | 13449 | -1.047 | -0.2593 | No | ||
| 108 | TNNC2 | 13496 | -1.056 | -0.2571 | No | ||
| 109 | RBMS3 | 13744 | -1.102 | -0.2656 | No | ||
| 110 | ITGA7 | 13878 | -1.127 | -0.2678 | No | ||
| 111 | BNIP3 | 13886 | -1.129 | -0.2632 | No | ||
| 112 | HCFC1R1 | 13998 | -1.152 | -0.2641 | No | ||
| 113 | CKM | 14402 | -1.244 | -0.2804 | Yes | ||
| 114 | LRRC20 | 14453 | -1.253 | -0.2776 | Yes | ||
| 115 | INPP4A | 14515 | -1.269 | -0.2753 | Yes | ||
| 116 | TCEA3 | 14556 | -1.276 | -0.2718 | Yes | ||
| 117 | CRTAP | 14659 | -1.296 | -0.2715 | Yes | ||
| 118 | DMD | 14733 | -1.309 | -0.2697 | Yes | ||
| 119 | EXTL1 | 14860 | -1.338 | -0.2706 | Yes | ||
| 120 | ADAM23 | 14893 | -1.345 | -0.2663 | Yes | ||
| 121 | NR4A1 | 14996 | -1.372 | -0.2658 | Yes | ||
| 122 | TYRO3 | 15081 | -1.390 | -0.2642 | Yes | ||
| 123 | SMPX | 15098 | -1.394 | -0.2589 | Yes | ||
| 124 | MYOZ2 | 15116 | -1.398 | -0.2536 | Yes | ||
| 125 | EHD1 | 15144 | -1.404 | -0.2488 | Yes | ||
| 126 | ENO3 | 15179 | -1.410 | -0.2444 | Yes | ||
| 127 | SOX5 | 15391 | -1.461 | -0.2494 | Yes | ||
| 128 | LIF | 15622 | -1.523 | -0.2551 | Yes | ||
| 129 | ARR3 | 15731 | -1.550 | -0.2540 | Yes | ||
| 130 | LRRTM4 | 15741 | -1.554 | -0.2476 | Yes | ||
| 131 | SLCO3A1 | 15770 | -1.564 | -0.2422 | Yes | ||
| 132 | RICS | 15870 | -1.592 | -0.2405 | Yes | ||
| 133 | SMYD1 | 15925 | -1.607 | -0.2363 | Yes | ||
| 134 | PTPRO | 15935 | -1.610 | -0.2297 | Yes | ||
| 135 | CYP2E1 | 16009 | -1.633 | -0.2264 | Yes | ||
| 136 | HFE2 | 16057 | -1.645 | -0.2216 | Yes | ||
| 137 | DLX5 | 16101 | -1.658 | -0.2166 | Yes | ||
| 138 | CAPN3 | 16137 | -1.666 | -0.2111 | Yes | ||
| 139 | HIST1H2BA | 16142 | -1.667 | -0.2039 | Yes | ||
| 140 | NR2F1 | 16196 | -1.681 | -0.1993 | Yes | ||
| 141 | POU4F1 | 16280 | -1.704 | -0.1963 | Yes | ||
| 142 | CLCN1 | 16286 | -1.705 | -0.1890 | Yes | ||
| 143 | BRUNOL4 | 16292 | -1.707 | -0.1817 | Yes | ||
| 144 | MAB21L2 | 16351 | -1.726 | -0.1772 | Yes | ||
| 145 | PCDH11X | 16399 | -1.744 | -0.1720 | Yes | ||
| 146 | TNFRSF17 | 16494 | -1.778 | -0.1692 | Yes | ||
| 147 | CACNA2D3 | 16500 | -1.782 | -0.1615 | Yes | ||
| 148 | USP2 | 16532 | -1.793 | -0.1553 | Yes | ||
| 149 | EEF1A2 | 16774 | -1.888 | -0.1600 | Yes | ||
| 150 | RASGRF1 | 16798 | -1.900 | -0.1528 | Yes | ||
| 151 | TTYH2 | 17157 | -2.038 | -0.1631 | Yes | ||
| 152 | KTN1 | 17230 | -2.074 | -0.1578 | Yes | ||
| 153 | SLC26A9 | 17280 | -2.095 | -0.1512 | Yes | ||
| 154 | ALS2CR2 | 17415 | -2.166 | -0.1489 | Yes | ||
| 155 | PRKAG1 | 17581 | -2.261 | -0.1478 | Yes | ||
| 156 | ADCY2 | 17595 | -2.269 | -0.1384 | Yes | ||
| 157 | CLDN23 | 17600 | -2.271 | -0.1285 | Yes | ||
| 158 | TWIST1 | 17639 | -2.293 | -0.1204 | Yes | ||
| 159 | PTPN1 | 17641 | -2.295 | -0.1103 | Yes | ||
| 160 | ELMO1 | 17910 | -2.505 | -0.1137 | Yes | ||
| 161 | HAS2 | 17943 | -2.542 | -0.1042 | Yes | ||
| 162 | PDLIM1 | 18174 | -2.796 | -0.1042 | Yes | ||
| 163 | MAP2K5 | 18221 | -2.871 | -0.0940 | Yes | ||
| 164 | RGS3 | 18334 | -3.081 | -0.0864 | Yes | ||
| 165 | NRXN3 | 18376 | -3.171 | -0.0745 | Yes | ||
| 166 | DTNB | 18478 | -3.435 | -0.0648 | Yes | ||
| 167 | RASGEF1B | 18493 | -3.482 | -0.0501 | Yes | ||
| 168 | TIMP2 | 18496 | -3.497 | -0.0347 | Yes | ||
| 169 | ATP2A3 | 18568 | -4.113 | -0.0203 | Yes | ||
| 170 | CSRP3 | 18610 | -5.144 | 0.0003 | Yes |
