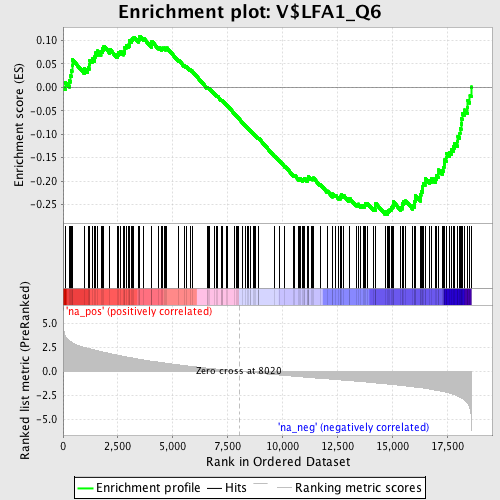
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$LFA1_Q6 |
| Enrichment Score (ES) | -0.27120072 |
| Normalized Enrichment Score (NES) | -1.3762777 |
| Nominal p-value | 0.014256619 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | M6PR | 96 | 3.705 | 0.0108 | No | ||
| 2 | IMMT | 285 | 3.171 | 0.0142 | No | ||
| 3 | BCL6 | 341 | 3.097 | 0.0246 | No | ||
| 4 | UPF2 | 374 | 3.039 | 0.0360 | No | ||
| 5 | NFAT5 | 408 | 2.982 | 0.0471 | No | ||
| 6 | EMILIN1 | 418 | 2.972 | 0.0594 | No | ||
| 7 | HOXC6 | 971 | 2.503 | 0.0402 | No | ||
| 8 | PYGO2 | 1134 | 2.399 | 0.0418 | No | ||
| 9 | CCNL1 | 1188 | 2.365 | 0.0491 | No | ||
| 10 | NXF1 | 1215 | 2.348 | 0.0578 | No | ||
| 11 | FLT1 | 1323 | 2.287 | 0.0619 | No | ||
| 12 | RBM12 | 1440 | 2.216 | 0.0652 | No | ||
| 13 | ARID1A | 1460 | 2.208 | 0.0736 | No | ||
| 14 | RBBP6 | 1554 | 2.151 | 0.0779 | No | ||
| 15 | PAX7 | 1727 | 2.062 | 0.0774 | No | ||
| 16 | UBL3 | 1791 | 2.031 | 0.0828 | No | ||
| 17 | BCL11B | 1857 | 1.998 | 0.0879 | No | ||
| 18 | ZFPM1 | 2136 | 1.843 | 0.0807 | No | ||
| 19 | CPNE1 | 2467 | 1.691 | 0.0701 | No | ||
| 20 | PNOC | 2514 | 1.675 | 0.0749 | No | ||
| 21 | FLI1 | 2608 | 1.630 | 0.0769 | No | ||
| 22 | KLK9 | 2754 | 1.564 | 0.0757 | No | ||
| 23 | LRP8 | 2807 | 1.536 | 0.0795 | No | ||
| 24 | GNAO1 | 2811 | 1.535 | 0.0860 | No | ||
| 25 | CASQ2 | 2869 | 1.511 | 0.0894 | No | ||
| 26 | PRRX1 | 2967 | 1.465 | 0.0905 | No | ||
| 27 | NACA | 3027 | 1.443 | 0.0935 | No | ||
| 28 | OBSCN | 3032 | 1.439 | 0.0995 | No | ||
| 29 | DHH | 3111 | 1.407 | 0.1013 | No | ||
| 30 | DLX1 | 3150 | 1.391 | 0.1053 | No | ||
| 31 | GSK3B | 3225 | 1.361 | 0.1071 | No | ||
| 32 | NPC2 | 3423 | 1.283 | 0.1020 | No | ||
| 33 | PHF12 | 3462 | 1.268 | 0.1054 | No | ||
| 34 | IL1F10 | 3490 | 1.259 | 0.1093 | No | ||
| 35 | NGFB | 3678 | 1.182 | 0.1043 | No | ||
| 36 | FXYD3 | 4025 | 1.055 | 0.0901 | No | ||
| 37 | ELF4 | 4026 | 1.055 | 0.0946 | No | ||
| 38 | PCBP2 | 4040 | 1.051 | 0.0985 | No | ||
| 39 | ARID4A | 4351 | 0.953 | 0.0858 | No | ||
| 40 | CXCR4 | 4476 | 0.915 | 0.0830 | No | ||
| 41 | BACE1 | 4528 | 0.897 | 0.0841 | No | ||
| 42 | NKX2-3 | 4598 | 0.872 | 0.0841 | No | ||
| 43 | CACNB1 | 4684 | 0.843 | 0.0831 | No | ||
| 44 | NPAS3 | 4732 | 0.831 | 0.0842 | No | ||
| 45 | LZTS2 | 5270 | 0.677 | 0.0580 | No | ||
| 46 | TAL1 | 5513 | 0.604 | 0.0475 | No | ||
| 47 | SIX4 | 5611 | 0.576 | 0.0447 | No | ||
| 48 | FKBP2 | 5786 | 0.528 | 0.0375 | No | ||
| 49 | TSP50 | 5874 | 0.507 | 0.0350 | No | ||
| 50 | LIN28 | 6572 | 0.335 | -0.0014 | No | ||
| 51 | NPTXR | 6587 | 0.332 | -0.0007 | No | ||
| 52 | S100A5 | 6630 | 0.321 | -0.0016 | No | ||
| 53 | BRPF1 | 6686 | 0.306 | -0.0033 | No | ||
| 54 | THRAP1 | 6877 | 0.262 | -0.0124 | No | ||
| 55 | SLC2A4 | 6993 | 0.234 | -0.0177 | No | ||
| 56 | OLFML2A | 7035 | 0.224 | -0.0189 | No | ||
| 57 | NEUROG1 | 7204 | 0.188 | -0.0272 | No | ||
| 58 | NYX | 7221 | 0.183 | -0.0273 | No | ||
| 59 | SP7 | 7249 | 0.176 | -0.0280 | No | ||
| 60 | RAB5C | 7272 | 0.170 | -0.0284 | No | ||
| 61 | CLTC | 7450 | 0.132 | -0.0375 | No | ||
| 62 | GBA2 | 7502 | 0.116 | -0.0397 | No | ||
| 63 | CKB | 7800 | 0.055 | -0.0556 | No | ||
| 64 | PLA2G10 | 7912 | 0.028 | -0.0615 | No | ||
| 65 | CRMP1 | 7965 | 0.015 | -0.0643 | No | ||
| 66 | SLC7A10 | 8008 | 0.003 | -0.0665 | No | ||
| 67 | MGEA5 | 8183 | -0.034 | -0.0758 | No | ||
| 68 | EDG1 | 8326 | -0.068 | -0.0832 | No | ||
| 69 | SMARCD2 | 8382 | -0.079 | -0.0859 | No | ||
| 70 | OTOP2 | 8448 | -0.093 | -0.0890 | No | ||
| 71 | CAV1 | 8525 | -0.112 | -0.0926 | No | ||
| 72 | PPY | 8558 | -0.117 | -0.0939 | No | ||
| 73 | MSX1 | 8669 | -0.138 | -0.0992 | No | ||
| 74 | CHX10 | 8724 | -0.149 | -0.1015 | No | ||
| 75 | LMOD3 | 8751 | -0.155 | -0.1023 | No | ||
| 76 | DUOX2 | 8896 | -0.185 | -0.1093 | No | ||
| 77 | NR5A2 | 8909 | -0.187 | -0.1091 | No | ||
| 78 | BOC | 8925 | -0.191 | -0.1091 | No | ||
| 79 | XYLT1 | 9644 | -0.350 | -0.1465 | No | ||
| 80 | GPR4 | 9845 | -0.386 | -0.1557 | No | ||
| 81 | STOML2 | 10111 | -0.434 | -0.1682 | No | ||
| 82 | RFX4 | 10515 | -0.511 | -0.1879 | No | ||
| 83 | ADAMTS4 | 10553 | -0.520 | -0.1876 | No | ||
| 84 | PAK1IP1 | 10746 | -0.554 | -0.1956 | No | ||
| 85 | DLGAP4 | 10793 | -0.563 | -0.1957 | No | ||
| 86 | MYBPH | 10819 | -0.567 | -0.1946 | No | ||
| 87 | WNT4 | 10925 | -0.588 | -0.1978 | No | ||
| 88 | USH1G | 10946 | -0.593 | -0.1963 | No | ||
| 89 | CTGF | 10982 | -0.599 | -0.1956 | No | ||
| 90 | KCNQ4 | 11009 | -0.604 | -0.1944 | No | ||
| 91 | GRIN2A | 11125 | -0.627 | -0.1980 | No | ||
| 92 | JPH4 | 11146 | -0.631 | -0.1963 | No | ||
| 93 | AKT3 | 11164 | -0.635 | -0.1945 | No | ||
| 94 | ADM | 11169 | -0.636 | -0.1920 | No | ||
| 95 | ACVR1C | 11174 | -0.638 | -0.1894 | No | ||
| 96 | UQCRC1 | 11318 | -0.665 | -0.1943 | No | ||
| 97 | SH3BP1 | 11381 | -0.673 | -0.1948 | No | ||
| 98 | SEMA6A | 11405 | -0.678 | -0.1931 | No | ||
| 99 | HIRA | 11708 | -0.727 | -0.2064 | No | ||
| 100 | LCAT | 12042 | -0.789 | -0.2210 | No | ||
| 101 | CIC | 12284 | -0.832 | -0.2305 | No | ||
| 102 | DPF2 | 12289 | -0.833 | -0.2271 | No | ||
| 103 | GPR3 | 12409 | -0.852 | -0.2299 | No | ||
| 104 | GABARAP | 12567 | -0.884 | -0.2346 | No | ||
| 105 | CREB3L2 | 12636 | -0.896 | -0.2344 | No | ||
| 106 | NFKBIA | 12659 | -0.899 | -0.2317 | No | ||
| 107 | BNC2 | 12666 | -0.900 | -0.2282 | No | ||
| 108 | NEURL | 12795 | -0.924 | -0.2311 | No | ||
| 109 | HOXC10 | 13030 | -0.966 | -0.2397 | No | ||
| 110 | CACNA1G | 13054 | -0.972 | -0.2367 | No | ||
| 111 | TRIP10 | 13361 | -1.032 | -0.2489 | No | ||
| 112 | ETV1 | 13444 | -1.046 | -0.2488 | No | ||
| 113 | NRGN | 13576 | -1.073 | -0.2513 | No | ||
| 114 | ZNF385 | 13686 | -1.092 | -0.2525 | No | ||
| 115 | SALL1 | 13747 | -1.103 | -0.2510 | No | ||
| 116 | KCNAB1 | 13763 | -1.106 | -0.2470 | No | ||
| 117 | RAB4B | 13866 | -1.126 | -0.2477 | No | ||
| 118 | PTRF | 14140 | -1.183 | -0.2574 | No | ||
| 119 | TNRC4 | 14234 | -1.203 | -0.2573 | No | ||
| 120 | MYOG | 14235 | -1.204 | -0.2521 | No | ||
| 121 | PPP1R16A | 14252 | -1.207 | -0.2477 | No | ||
| 122 | SESN3 | 14686 | -1.301 | -0.2656 | Yes | ||
| 123 | LAG3 | 14786 | -1.323 | -0.2653 | Yes | ||
| 124 | KIF1C | 14838 | -1.333 | -0.2623 | Yes | ||
| 125 | MRPL40 | 14895 | -1.346 | -0.2595 | Yes | ||
| 126 | HNRPR | 14949 | -1.360 | -0.2565 | Yes | ||
| 127 | NR4A1 | 14996 | -1.372 | -0.2531 | Yes | ||
| 128 | ACCN1 | 15042 | -1.381 | -0.2496 | Yes | ||
| 129 | EPHB2 | 15046 | -1.382 | -0.2438 | Yes | ||
| 130 | SOX5 | 15391 | -1.461 | -0.2561 | Yes | ||
| 131 | FES | 15456 | -1.479 | -0.2532 | Yes | ||
| 132 | RABGGTA | 15479 | -1.487 | -0.2480 | Yes | ||
| 133 | EPO | 15525 | -1.499 | -0.2440 | Yes | ||
| 134 | PALM | 15613 | -1.520 | -0.2421 | Yes | ||
| 135 | RASGRP2 | 15941 | -1.614 | -0.2529 | Yes | ||
| 136 | ANXA4 | 16007 | -1.633 | -0.2494 | Yes | ||
| 137 | GAD1 | 16028 | -1.639 | -0.2434 | Yes | ||
| 138 | TCF7 | 16050 | -1.644 | -0.2375 | Yes | ||
| 139 | GABRG2 | 16058 | -1.646 | -0.2308 | Yes | ||
| 140 | POU4F1 | 16280 | -1.704 | -0.2354 | Yes | ||
| 141 | SMAD3 | 16301 | -1.711 | -0.2291 | Yes | ||
| 142 | RHO | 16314 | -1.715 | -0.2224 | Yes | ||
| 143 | NDUFS2 | 16368 | -1.733 | -0.2178 | Yes | ||
| 144 | TBR1 | 16392 | -1.742 | -0.2115 | Yes | ||
| 145 | HEBP1 | 16402 | -1.745 | -0.2045 | Yes | ||
| 146 | COX7A2L | 16499 | -1.782 | -0.2020 | Yes | ||
| 147 | CLIC5 | 16507 | -1.785 | -0.1947 | Yes | ||
| 148 | SLC13A5 | 16719 | -1.862 | -0.1981 | Yes | ||
| 149 | ZFP36 | 16799 | -1.900 | -0.1942 | Yes | ||
| 150 | GSS | 16973 | -1.966 | -0.1951 | Yes | ||
| 151 | EFNA3 | 17007 | -1.979 | -0.1883 | Yes | ||
| 152 | SLC25A29 | 17097 | -2.018 | -0.1844 | Yes | ||
| 153 | CNN2 | 17103 | -2.020 | -0.1760 | Yes | ||
| 154 | SLC26A9 | 17280 | -2.095 | -0.1765 | Yes | ||
| 155 | TITF1 | 17338 | -2.121 | -0.1705 | Yes | ||
| 156 | CEBPB | 17365 | -2.138 | -0.1627 | Yes | ||
| 157 | CNKSR2 | 17390 | -2.150 | -0.1547 | Yes | ||
| 158 | MCRS1 | 17465 | -2.191 | -0.1493 | Yes | ||
| 159 | BAX | 17473 | -2.195 | -0.1402 | Yes | ||
| 160 | PLEC1 | 17623 | -2.286 | -0.1384 | Yes | ||
| 161 | PDLIM7 | 17707 | -2.343 | -0.1328 | Yes | ||
| 162 | DCTN1 | 17781 | -2.405 | -0.1264 | Yes | ||
| 163 | NCDN | 17843 | -2.457 | -0.1191 | Yes | ||
| 164 | MAZ | 17977 | -2.573 | -0.1152 | Yes | ||
| 165 | MNT | 17992 | -2.589 | -0.1048 | Yes | ||
| 166 | ROM1 | 18082 | -2.685 | -0.0981 | Yes | ||
| 167 | VASP | 18094 | -2.698 | -0.0870 | Yes | ||
| 168 | PDYN | 18140 | -2.755 | -0.0776 | Yes | ||
| 169 | TPCN1 | 18163 | -2.782 | -0.0668 | Yes | ||
| 170 | HES1 | 18189 | -2.814 | -0.0560 | Yes | ||
| 171 | RHOG | 18281 | -2.964 | -0.0482 | Yes | ||
| 172 | POLD4 | 18431 | -3.299 | -0.0420 | Yes | ||
| 173 | IFRD2 | 18445 | -3.338 | -0.0283 | Yes | ||
| 174 | HARS2 | 18544 | -3.849 | -0.0170 | Yes | ||
| 175 | MOV10 | 18600 | -4.845 | 0.0009 | Yes |
