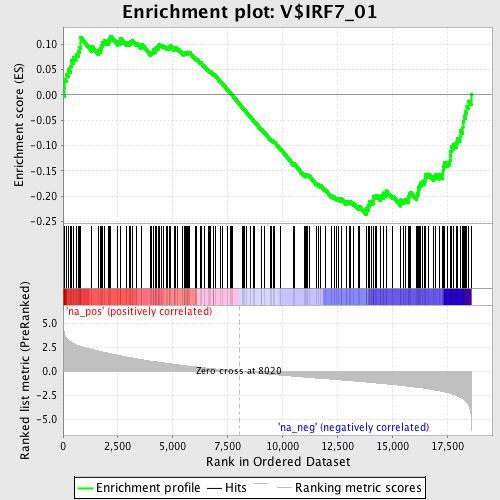
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$IRF7_01 |
| Enrichment Score (ES) | -0.23590267 |
| Normalized Enrichment Score (NES) | -1.1927094 |
| Nominal p-value | 0.11336032 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TAF5 | 50 | 4.062 | 0.0144 | No | ||
| 2 | CDYL | 64 | 3.934 | 0.0302 | No | ||
| 3 | VPS24 | 142 | 3.516 | 0.0408 | No | ||
| 4 | TRPM3 | 239 | 3.256 | 0.0492 | No | ||
| 5 | BCL6 | 341 | 3.097 | 0.0568 | No | ||
| 6 | UPF2 | 374 | 3.039 | 0.0678 | No | ||
| 7 | HTR5A | 482 | 2.901 | 0.0742 | No | ||
| 8 | ERG | 589 | 2.789 | 0.0801 | No | ||
| 9 | GNA13 | 685 | 2.706 | 0.0863 | No | ||
| 10 | PSMA3 | 751 | 2.659 | 0.0940 | No | ||
| 11 | TLK2 | 800 | 2.616 | 0.1024 | No | ||
| 12 | RBL1 | 801 | 2.613 | 0.1133 | No | ||
| 13 | PPP1R10 | 1312 | 2.293 | 0.0953 | No | ||
| 14 | TLX2 | 1607 | 2.124 | 0.0883 | No | ||
| 15 | NT5C3 | 1713 | 2.067 | 0.0913 | No | ||
| 16 | DAPP1 | 1760 | 2.045 | 0.0974 | No | ||
| 17 | PDE4D | 1800 | 2.027 | 0.1038 | No | ||
| 18 | TCF12 | 1894 | 1.970 | 0.1070 | No | ||
| 19 | YTHDF2 | 2054 | 1.887 | 0.1063 | No | ||
| 20 | LRP6 | 2124 | 1.852 | 0.1103 | No | ||
| 21 | IRF2 | 2168 | 1.830 | 0.1157 | No | ||
| 22 | ARRDC3 | 2496 | 1.680 | 0.1050 | No | ||
| 23 | FLI1 | 2608 | 1.630 | 0.1058 | No | ||
| 24 | SOX4 | 2626 | 1.618 | 0.1117 | No | ||
| 25 | USP18 | 2896 | 1.498 | 0.1034 | No | ||
| 26 | TUBD1 | 3029 | 1.441 | 0.1023 | No | ||
| 27 | IFIT2 | 3086 | 1.416 | 0.1052 | No | ||
| 28 | DLX1 | 3150 | 1.391 | 0.1077 | No | ||
| 29 | DDX58 | 3355 | 1.312 | 0.1021 | No | ||
| 30 | COMMD10 | 3550 | 1.232 | 0.0968 | No | ||
| 31 | SLC4A10 | 3592 | 1.215 | 0.0996 | No | ||
| 32 | CBX4 | 4001 | 1.065 | 0.0820 | No | ||
| 33 | BST2 | 4042 | 1.050 | 0.0842 | No | ||
| 34 | AAMP | 4111 | 1.027 | 0.0848 | No | ||
| 35 | FOXP2 | 4119 | 1.024 | 0.0888 | No | ||
| 36 | GATA6 | 4207 | 1.000 | 0.0882 | No | ||
| 37 | SST | 4223 | 0.995 | 0.0916 | No | ||
| 38 | PITX2 | 4263 | 0.981 | 0.0936 | No | ||
| 39 | PSME1 | 4330 | 0.960 | 0.0941 | No | ||
| 40 | PDE1B | 4332 | 0.959 | 0.0980 | No | ||
| 41 | MPL | 4384 | 0.944 | 0.0992 | No | ||
| 42 | CXCR4 | 4476 | 0.915 | 0.0982 | No | ||
| 43 | HIVEP1 | 4582 | 0.878 | 0.0961 | No | ||
| 44 | NFIX | 4696 | 0.840 | 0.0935 | No | ||
| 45 | NR4A3 | 4766 | 0.823 | 0.0933 | No | ||
| 46 | EPHA7 | 4832 | 0.807 | 0.0931 | No | ||
| 47 | HS6ST2 | 4849 | 0.803 | 0.0956 | No | ||
| 48 | ZFP36L1 | 4874 | 0.797 | 0.0977 | No | ||
| 49 | KIRREL3 | 5058 | 0.736 | 0.0908 | No | ||
| 50 | GSC | 5084 | 0.727 | 0.0925 | No | ||
| 51 | DICER1 | 5111 | 0.720 | 0.0941 | No | ||
| 52 | EYA1 | 5231 | 0.687 | 0.0906 | No | ||
| 53 | SIPA1L1 | 5450 | 0.620 | 0.0814 | No | ||
| 54 | RFX5 | 5509 | 0.605 | 0.0808 | No | ||
| 55 | TREX2 | 5521 | 0.602 | 0.0827 | No | ||
| 56 | FIBCD1 | 5568 | 0.588 | 0.0827 | No | ||
| 57 | PKN3 | 5615 | 0.575 | 0.0826 | No | ||
| 58 | E2F3 | 5681 | 0.554 | 0.0814 | No | ||
| 59 | POPDC2 | 5688 | 0.553 | 0.0834 | No | ||
| 60 | ZHX2 | 5732 | 0.541 | 0.0833 | No | ||
| 61 | PSCD3 | 5756 | 0.536 | 0.0843 | No | ||
| 62 | ARTN | 6037 | 0.465 | 0.0711 | No | ||
| 63 | IL22RA1 | 6089 | 0.455 | 0.0702 | No | ||
| 64 | ABI3 | 6247 | 0.412 | 0.0635 | No | ||
| 65 | KPNA3 | 6286 | 0.403 | 0.0631 | No | ||
| 66 | SLC25A28 | 6465 | 0.360 | 0.0549 | No | ||
| 67 | PRDM16 | 6648 | 0.317 | 0.0464 | No | ||
| 68 | NOG | 6679 | 0.308 | 0.0461 | No | ||
| 69 | PCBP1 | 6722 | 0.298 | 0.0450 | No | ||
| 70 | ELK4 | 6858 | 0.266 | 0.0388 | No | ||
| 71 | CACNA1A | 6869 | 0.263 | 0.0394 | No | ||
| 72 | NPR3 | 6872 | 0.262 | 0.0404 | No | ||
| 73 | BAT5 | 6950 | 0.244 | 0.0372 | No | ||
| 74 | MAP2K6 | 7150 | 0.203 | 0.0273 | No | ||
| 75 | TSC1 | 7262 | 0.172 | 0.0220 | No | ||
| 76 | ZDHHC14 | 7496 | 0.119 | 0.0099 | No | ||
| 77 | ZBTB10 | 7622 | 0.092 | 0.0035 | No | ||
| 78 | LHX9 | 7678 | 0.081 | 0.0008 | No | ||
| 79 | NOS1 | 7725 | 0.069 | -0.0014 | No | ||
| 80 | LOX | 8177 | -0.033 | -0.0257 | No | ||
| 81 | AMY2A | 8223 | -0.044 | -0.0280 | No | ||
| 82 | BMP5 | 8244 | -0.048 | -0.0288 | No | ||
| 83 | PSMB9 | 8339 | -0.070 | -0.0337 | No | ||
| 84 | OPTN | 8521 | -0.111 | -0.0430 | No | ||
| 85 | MSX1 | 8669 | -0.138 | -0.0504 | No | ||
| 86 | EGR4 | 8714 | -0.146 | -0.0522 | No | ||
| 87 | PLEK2 | 9035 | -0.216 | -0.0686 | No | ||
| 88 | ESR1 | 9054 | -0.220 | -0.0687 | No | ||
| 89 | PLXNC1 | 9171 | -0.245 | -0.0739 | No | ||
| 90 | MAP3K8 | 9464 | -0.310 | -0.0885 | No | ||
| 91 | CCL3 | 9517 | -0.323 | -0.0899 | No | ||
| 92 | LHX5 | 9594 | -0.339 | -0.0926 | No | ||
| 93 | CXCL10 | 9639 | -0.349 | -0.0936 | No | ||
| 94 | HMGB3 | 9923 | -0.400 | -0.1072 | No | ||
| 95 | RFX4 | 10515 | -0.511 | -0.1371 | No | ||
| 96 | FOS | 10529 | -0.514 | -0.1357 | No | ||
| 97 | COL12A1 | 10997 | -0.602 | -0.1585 | No | ||
| 98 | TIA1 | 11063 | -0.614 | -0.1594 | No | ||
| 99 | SOCS1 | 11071 | -0.617 | -0.1572 | No | ||
| 100 | JPH4 | 11146 | -0.631 | -0.1586 | No | ||
| 101 | EN1 | 11217 | -0.645 | -0.1597 | No | ||
| 102 | DLX4 | 11546 | -0.703 | -0.1745 | No | ||
| 103 | DLL4 | 11649 | -0.719 | -0.1770 | No | ||
| 104 | EGFL6 | 11748 | -0.732 | -0.1793 | No | ||
| 105 | HOXB3 | 11961 | -0.773 | -0.1875 | No | ||
| 106 | GDNF | 12227 | -0.822 | -0.1984 | No | ||
| 107 | IFNB1 | 12347 | -0.842 | -0.2014 | No | ||
| 108 | LHX6 | 12459 | -0.863 | -0.2038 | No | ||
| 109 | IL27 | 12538 | -0.877 | -0.2043 | No | ||
| 110 | BNC2 | 12666 | -0.900 | -0.2074 | No | ||
| 111 | KIF13A | 12695 | -0.905 | -0.2051 | No | ||
| 112 | SLC12A7 | 12894 | -0.944 | -0.2119 | No | ||
| 113 | PDGFC | 12935 | -0.951 | -0.2101 | No | ||
| 114 | HOXC10 | 13030 | -0.966 | -0.2111 | No | ||
| 115 | HOXD13 | 13080 | -0.976 | -0.2097 | No | ||
| 116 | A2BP1 | 13231 | -1.005 | -0.2136 | No | ||
| 117 | CPT1A | 13467 | -1.052 | -0.2219 | No | ||
| 118 | GRM3 | 13518 | -1.063 | -0.2202 | No | ||
| 119 | PRDM13 | 13809 | -1.115 | -0.2312 | Yes | ||
| 120 | TCF15 | 13819 | -1.116 | -0.2270 | Yes | ||
| 121 | SP110 | 13834 | -1.119 | -0.2231 | Yes | ||
| 122 | TNFSF15 | 13921 | -1.138 | -0.2230 | Yes | ||
| 123 | ITGB7 | 13931 | -1.140 | -0.2187 | Yes | ||
| 124 | STX16 | 13947 | -1.142 | -0.2147 | Yes | ||
| 125 | CUL2 | 13969 | -1.147 | -0.2110 | Yes | ||
| 126 | IL4 | 14062 | -1.165 | -0.2111 | Yes | ||
| 127 | STX11 | 14134 | -1.182 | -0.2100 | Yes | ||
| 128 | FLRT2 | 14141 | -1.183 | -0.2053 | Yes | ||
| 129 | ELAVL2 | 14146 | -1.184 | -0.2006 | Yes | ||
| 130 | PABPC4 | 14220 | -1.200 | -0.1995 | Yes | ||
| 131 | NR4A2 | 14288 | -1.217 | -0.1980 | Yes | ||
| 132 | FEV | 14485 | -1.261 | -0.2034 | Yes | ||
| 133 | IL18BP | 14487 | -1.262 | -0.1981 | Yes | ||
| 134 | APOBEC2 | 14596 | -1.284 | -0.1986 | Yes | ||
| 135 | RDH10 | 14602 | -1.285 | -0.1935 | Yes | ||
| 136 | GDI1 | 14718 | -1.305 | -0.1942 | Yes | ||
| 137 | DMD | 14733 | -1.309 | -0.1895 | Yes | ||
| 138 | FHL3 | 15027 | -1.378 | -0.1996 | Yes | ||
| 139 | SOX5 | 15391 | -1.461 | -0.2131 | Yes | ||
| 140 | HOXA10 | 15394 | -1.462 | -0.2071 | Yes | ||
| 141 | GPX1 | 15529 | -1.500 | -0.2081 | Yes | ||
| 142 | LIF | 15622 | -1.523 | -0.2067 | Yes | ||
| 143 | LZIC | 15735 | -1.552 | -0.2062 | Yes | ||
| 144 | LRRTM4 | 15741 | -1.554 | -0.2000 | Yes | ||
| 145 | VIT | 15772 | -1.565 | -0.1950 | Yes | ||
| 146 | ADAR | 15835 | -1.582 | -0.1917 | Yes | ||
| 147 | ECEL1 | 16115 | -1.662 | -0.1999 | Yes | ||
| 148 | LDB2 | 16135 | -1.666 | -0.1939 | Yes | ||
| 149 | MXD3 | 16180 | -1.676 | -0.1893 | Yes | ||
| 150 | CALD1 | 16193 | -1.681 | -0.1829 | Yes | ||
| 151 | ATP5G2 | 16259 | -1.698 | -0.1792 | Yes | ||
| 152 | BRUNOL4 | 16292 | -1.707 | -0.1738 | Yes | ||
| 153 | HOXB8 | 16370 | -1.734 | -0.1707 | Yes | ||
| 154 | RPS6KB1 | 16473 | -1.770 | -0.1688 | Yes | ||
| 155 | CACNA2D3 | 16500 | -1.782 | -0.1627 | Yes | ||
| 156 | USP2 | 16532 | -1.793 | -0.1569 | Yes | ||
| 157 | CHST8 | 16661 | -1.840 | -0.1561 | Yes | ||
| 158 | POLL | 16892 | -1.934 | -0.1604 | Yes | ||
| 159 | PPP2R2A | 16989 | -1.973 | -0.1574 | Yes | ||
| 160 | COL11A2 | 17151 | -2.036 | -0.1576 | Yes | ||
| 161 | RFX3 | 17292 | -2.101 | -0.1563 | Yes | ||
| 162 | RAPH1 | 17314 | -2.109 | -0.1486 | Yes | ||
| 163 | CASP7 | 17336 | -2.120 | -0.1408 | Yes | ||
| 164 | GJC1 | 17367 | -2.138 | -0.1335 | Yes | ||
| 165 | GNGT2 | 17523 | -2.229 | -0.1325 | Yes | ||
| 166 | TAP1 | 17633 | -2.292 | -0.1288 | Yes | ||
| 167 | ANGPTL4 | 17656 | -2.310 | -0.1203 | Yes | ||
| 168 | ERBB2 | 17667 | -2.319 | -0.1111 | Yes | ||
| 169 | GRM7 | 17692 | -2.335 | -0.1026 | Yes | ||
| 170 | TAPBP | 17775 | -2.399 | -0.0970 | Yes | ||
| 171 | ARF6 | 17916 | -2.511 | -0.0940 | Yes | ||
| 172 | PIK3CD | 17957 | -2.549 | -0.0855 | Yes | ||
| 173 | MRPS18B | 18090 | -2.693 | -0.0813 | Yes | ||
| 174 | CXCL16 | 18097 | -2.706 | -0.0703 | Yes | ||
| 175 | MAP2K5 | 18221 | -2.871 | -0.0649 | Yes | ||
| 176 | EVL | 18227 | -2.892 | -0.0530 | Yes | ||
| 177 | LGALS3BP | 18271 | -2.951 | -0.0430 | Yes | ||
| 178 | LAPTM4A | 18341 | -3.096 | -0.0337 | Yes | ||
| 179 | AGPAT4 | 18406 | -3.238 | -0.0236 | Yes | ||
| 180 | UBE1L | 18480 | -3.449 | -0.0130 | Yes | ||
| 181 | MOV10 | 18600 | -4.845 | 0.0009 | Yes |
