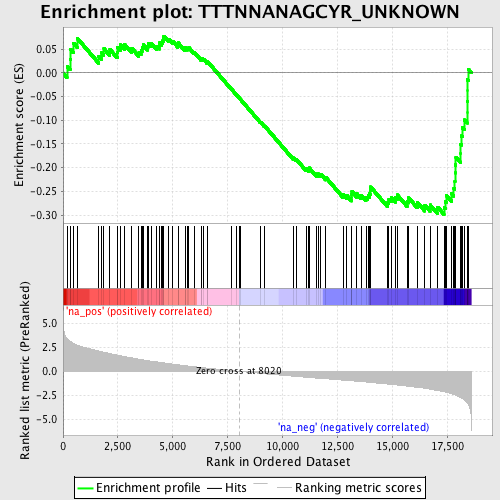
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTTNNANAGCYR_UNKNOWN |
| Enrichment Score (ES) | -0.29885656 |
| Normalized Enrichment Score (NES) | -1.3644663 |
| Nominal p-value | 0.039447732 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HOXB4 | 180 | 3.412 | 0.0144 | No | ||
| 2 | ACN9 | 321 | 3.125 | 0.0289 | No | ||
| 3 | GRK5 | 343 | 3.087 | 0.0496 | No | ||
| 4 | NDRG3 | 477 | 2.908 | 0.0630 | No | ||
| 5 | RNF44 | 660 | 2.727 | 0.0725 | No | ||
| 6 | CHD4 | 1626 | 2.116 | 0.0353 | No | ||
| 7 | NFATC1 | 1761 | 2.044 | 0.0426 | No | ||
| 8 | AMOT | 1860 | 1.994 | 0.0514 | No | ||
| 9 | CYP51A1 | 2132 | 1.847 | 0.0498 | No | ||
| 10 | CPNE1 | 2467 | 1.691 | 0.0437 | No | ||
| 11 | MXI1 | 2486 | 1.684 | 0.0547 | No | ||
| 12 | SLC6A13 | 2612 | 1.629 | 0.0594 | No | ||
| 13 | SLC10A2 | 2806 | 1.536 | 0.0599 | No | ||
| 14 | HIST1H1D | 3130 | 1.401 | 0.0523 | No | ||
| 15 | HIST3H2A | 3456 | 1.272 | 0.0438 | No | ||
| 16 | AOC2 | 3555 | 1.230 | 0.0472 | No | ||
| 17 | USP47 | 3601 | 1.213 | 0.0533 | No | ||
| 18 | TCF4 | 3643 | 1.198 | 0.0596 | No | ||
| 19 | CAST | 3855 | 1.120 | 0.0561 | No | ||
| 20 | RANGAP1 | 3882 | 1.109 | 0.0626 | No | ||
| 21 | PRKCQ | 4013 | 1.060 | 0.0630 | No | ||
| 22 | SLC6A1 | 4259 | 0.983 | 0.0568 | No | ||
| 23 | HIST1H2BE | 4377 | 0.945 | 0.0571 | No | ||
| 24 | AGTR2 | 4382 | 0.944 | 0.0636 | No | ||
| 25 | DGKG | 4492 | 0.910 | 0.0641 | No | ||
| 26 | DSC1 | 4543 | 0.891 | 0.0677 | No | ||
| 27 | FGF17 | 4563 | 0.883 | 0.0729 | No | ||
| 28 | OTP | 4589 | 0.876 | 0.0778 | No | ||
| 29 | STARD13 | 4815 | 0.811 | 0.0714 | No | ||
| 30 | HOXB6 | 4991 | 0.760 | 0.0673 | No | ||
| 31 | SLC12A2 | 5236 | 0.685 | 0.0590 | No | ||
| 32 | CENTD1 | 5239 | 0.685 | 0.0637 | No | ||
| 33 | HIST1H2AA | 5560 | 0.592 | 0.0506 | No | ||
| 34 | PPP3CA | 5567 | 0.589 | 0.0545 | No | ||
| 35 | E2F3 | 5681 | 0.554 | 0.0523 | No | ||
| 36 | CLDN2 | 5731 | 0.541 | 0.0535 | No | ||
| 37 | FOXP1 | 5973 | 0.483 | 0.0439 | No | ||
| 38 | NRAP | 6308 | 0.397 | 0.0286 | No | ||
| 39 | DHRS3 | 6324 | 0.394 | 0.0306 | No | ||
| 40 | HIST1H1C | 6411 | 0.371 | 0.0286 | No | ||
| 41 | LIN28 | 6572 | 0.335 | 0.0223 | No | ||
| 42 | HIST2H2AC | 6583 | 0.332 | 0.0241 | No | ||
| 43 | HRASLS | 7685 | 0.079 | -0.0348 | No | ||
| 44 | SPRED1 | 7924 | 0.025 | -0.0474 | No | ||
| 45 | HIST3H2BB | 8042 | -0.003 | -0.0537 | No | ||
| 46 | HIST1H2AH | 8089 | -0.016 | -0.0561 | No | ||
| 47 | KCNIP4 | 9000 | -0.207 | -0.1038 | No | ||
| 48 | NDNL2 | 9182 | -0.246 | -0.1118 | No | ||
| 49 | NAP1L5 | 10499 | -0.508 | -0.1793 | No | ||
| 50 | HIST1H2AC | 10622 | -0.532 | -0.1821 | No | ||
| 51 | CNN1 | 11078 | -0.619 | -0.2023 | No | ||
| 52 | SERPINA7 | 11181 | -0.639 | -0.2033 | No | ||
| 53 | KCNK12 | 11212 | -0.645 | -0.2004 | No | ||
| 54 | GPM6B | 11543 | -0.702 | -0.2132 | No | ||
| 55 | DNAJB8 | 11636 | -0.718 | -0.2131 | No | ||
| 56 | TCERG1L | 11741 | -0.731 | -0.2136 | No | ||
| 57 | HIST1H2AI | 11973 | -0.775 | -0.2206 | No | ||
| 58 | CDH16 | 12766 | -0.920 | -0.2568 | No | ||
| 59 | MEIS1 | 12937 | -0.952 | -0.2593 | No | ||
| 60 | MYH2 | 13144 | -0.988 | -0.2634 | No | ||
| 61 | ONECUT2 | 13148 | -0.988 | -0.2566 | No | ||
| 62 | BMPR2 | 13162 | -0.992 | -0.2503 | No | ||
| 63 | HIST1H1T | 13386 | -1.036 | -0.2550 | No | ||
| 64 | MTSS1 | 13588 | -1.076 | -0.2582 | No | ||
| 65 | FGD4 | 13806 | -1.114 | -0.2621 | No | ||
| 66 | TBXAS1 | 13896 | -1.132 | -0.2589 | No | ||
| 67 | HIST1H2BC | 13977 | -1.148 | -0.2551 | No | ||
| 68 | OLR1 | 14003 | -1.153 | -0.2483 | No | ||
| 69 | SMO | 14014 | -1.155 | -0.2407 | No | ||
| 70 | ETS1 | 14799 | -1.325 | -0.2736 | No | ||
| 71 | LHX2 | 14847 | -1.335 | -0.2667 | No | ||
| 72 | BLNK | 14954 | -1.362 | -0.2628 | No | ||
| 73 | HIST1H3F | 15141 | -1.403 | -0.2629 | No | ||
| 74 | HIST1H1E | 15226 | -1.421 | -0.2574 | No | ||
| 75 | INPPL1 | 15679 | -1.538 | -0.2710 | No | ||
| 76 | RGN | 15748 | -1.556 | -0.2636 | No | ||
| 77 | HIST1H2BA | 16142 | -1.667 | -0.2731 | No | ||
| 78 | HIST1H2AE | 16487 | -1.775 | -0.2791 | No | ||
| 79 | SLC35A2 | 16724 | -1.864 | -0.2786 | No | ||
| 80 | CLDN10 | 17072 | -2.005 | -0.2832 | No | ||
| 81 | HIST1H4A | 17363 | -2.136 | -0.2837 | Yes | ||
| 82 | HIST1H2BG | 17426 | -2.169 | -0.2718 | Yes | ||
| 83 | HIST1H2BK | 17462 | -2.189 | -0.2582 | Yes | ||
| 84 | FBXO21 | 17709 | -2.345 | -0.2549 | Yes | ||
| 85 | HIST1H2BB | 17808 | -2.429 | -0.2430 | Yes | ||
| 86 | DPT | 17857 | -2.464 | -0.2282 | Yes | ||
| 87 | HIST1H2BJ | 17864 | -2.467 | -0.2110 | Yes | ||
| 88 | HIST1H2BN | 17871 | -2.472 | -0.1939 | Yes | ||
| 89 | HIST1H2BF | 17902 | -2.498 | -0.1778 | Yes | ||
| 90 | HIST1H1B | 18117 | -2.734 | -0.1701 | Yes | ||
| 91 | HIST1H2BL | 18122 | -2.737 | -0.1509 | Yes | ||
| 92 | HIST1H2BH | 18151 | -2.770 | -0.1328 | Yes | ||
| 93 | HIST1H3H | 18193 | -2.820 | -0.1151 | Yes | ||
| 94 | HIST1H3B | 18287 | -2.978 | -0.0991 | Yes | ||
| 95 | HIST2H2BE | 18414 | -3.254 | -0.0829 | Yes | ||
| 96 | HIST1H3D | 18416 | -3.261 | -0.0599 | Yes | ||
| 97 | HIST1H3C | 18426 | -3.285 | -0.0371 | Yes | ||
| 98 | HIST1H1A | 18428 | -3.292 | -0.0139 | Yes | ||
| 99 | HIST1H2AB | 18468 | -3.391 | 0.0080 | Yes |
