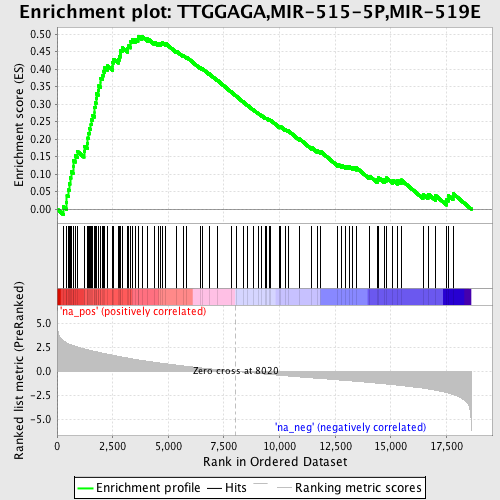
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TTGGAGA,MIR-515-5P,MIR-519E |
| Enrichment Score (ES) | 0.49420205 |
| Normalized Enrichment Score (NES) | 2.251911 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.5224124E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SFRS1 | 283 | 3.183 | 0.0067 | Yes | ||
| 2 | RBL2 | 413 | 2.976 | 0.0204 | Yes | ||
| 3 | RIOK3 | 443 | 2.943 | 0.0392 | Yes | ||
| 4 | AP3S1 | 517 | 2.860 | 0.0550 | Yes | ||
| 5 | FRAS1 | 544 | 2.831 | 0.0732 | Yes | ||
| 6 | MARCKS | 580 | 2.799 | 0.0907 | Yes | ||
| 7 | RPP14 | 625 | 2.757 | 0.1074 | Yes | ||
| 8 | MTHFD1L | 721 | 2.683 | 0.1208 | Yes | ||
| 9 | KPNA1 | 728 | 2.676 | 0.1390 | Yes | ||
| 10 | EPC2 | 813 | 2.604 | 0.1525 | Yes | ||
| 11 | ZBTB1 | 922 | 2.534 | 0.1642 | Yes | ||
| 12 | TMEM2 | 1209 | 2.352 | 0.1650 | Yes | ||
| 13 | CYR61 | 1235 | 2.338 | 0.1798 | Yes | ||
| 14 | ATXN3 | 1364 | 2.261 | 0.1886 | Yes | ||
| 15 | ATRN | 1382 | 2.250 | 0.2032 | Yes | ||
| 16 | RAP2C | 1416 | 2.232 | 0.2169 | Yes | ||
| 17 | ARID1A | 1460 | 2.208 | 0.2299 | Yes | ||
| 18 | MAPRE1 | 1521 | 2.168 | 0.2416 | Yes | ||
| 19 | DONSON | 1545 | 2.156 | 0.2553 | Yes | ||
| 20 | UBAP2 | 1578 | 2.141 | 0.2684 | Yes | ||
| 21 | DDX3Y | 1680 | 2.085 | 0.2774 | Yes | ||
| 22 | SLC9A2 | 1683 | 2.084 | 0.2917 | Yes | ||
| 23 | NOTCH2 | 1715 | 2.067 | 0.3043 | Yes | ||
| 24 | HIP2 | 1753 | 2.053 | 0.3165 | Yes | ||
| 25 | NCOA1 | 1776 | 2.036 | 0.3294 | Yes | ||
| 26 | UHRF2 | 1844 | 2.003 | 0.3397 | Yes | ||
| 27 | RAP1B | 1871 | 1.987 | 0.3520 | Yes | ||
| 28 | HNRPUL1 | 1959 | 1.935 | 0.3607 | Yes | ||
| 29 | ICK | 1960 | 1.935 | 0.3741 | Yes | ||
| 30 | YTHDF2 | 2054 | 1.887 | 0.3821 | Yes | ||
| 31 | GABBR1 | 2090 | 1.869 | 0.3932 | Yes | ||
| 32 | RDH11 | 2134 | 1.844 | 0.4036 | Yes | ||
| 33 | RSBN1 | 2256 | 1.791 | 0.4095 | Yes | ||
| 34 | DDX3X | 2492 | 1.681 | 0.4084 | Yes | ||
| 35 | ARRDC3 | 2496 | 1.680 | 0.4199 | Yes | ||
| 36 | ULK2 | 2547 | 1.657 | 0.4286 | Yes | ||
| 37 | RPS6KA3 | 2760 | 1.562 | 0.4280 | Yes | ||
| 38 | MOSPD2 | 2804 | 1.537 | 0.4363 | Yes | ||
| 39 | PUM1 | 2840 | 1.521 | 0.4449 | Yes | ||
| 40 | SUMO2 | 2858 | 1.516 | 0.4545 | Yes | ||
| 41 | IQGAP1 | 2923 | 1.489 | 0.4614 | Yes | ||
| 42 | YAF2 | 3163 | 1.388 | 0.4581 | Yes | ||
| 43 | POU2F1 | 3186 | 1.374 | 0.4664 | Yes | ||
| 44 | UBE2H | 3301 | 1.330 | 0.4694 | Yes | ||
| 45 | PLAGL2 | 3302 | 1.330 | 0.4786 | Yes | ||
| 46 | CMTM6 | 3372 | 1.305 | 0.4839 | Yes | ||
| 47 | ARHGEF3 | 3509 | 1.252 | 0.4853 | Yes | ||
| 48 | CAB39 | 3636 | 1.201 | 0.4868 | Yes | ||
| 49 | EPN2 | 3652 | 1.193 | 0.4942 | Yes | ||
| 50 | SOX12 | 3816 | 1.134 | 0.4932 | No | ||
| 51 | PSMD11 | 4075 | 1.037 | 0.4865 | No | ||
| 52 | SLC7A11 | 4396 | 0.939 | 0.4757 | No | ||
| 53 | POM121 | 4554 | 0.886 | 0.4734 | No | ||
| 54 | KCTD7 | 4659 | 0.850 | 0.4736 | No | ||
| 55 | BSN | 4717 | 0.834 | 0.4763 | No | ||
| 56 | ZFP36L1 | 4874 | 0.797 | 0.4734 | No | ||
| 57 | MTF2 | 5367 | 0.642 | 0.4513 | No | ||
| 58 | UBE2B | 5673 | 0.557 | 0.4387 | No | ||
| 59 | EMX2 | 5833 | 0.515 | 0.4336 | No | ||
| 60 | SCN8A | 6433 | 0.367 | 0.4038 | No | ||
| 61 | OPHN1 | 6523 | 0.345 | 0.4014 | No | ||
| 62 | ETNK2 | 6830 | 0.272 | 0.3868 | No | ||
| 63 | ANUBL1 | 7206 | 0.188 | 0.3678 | No | ||
| 64 | BTG2 | 7819 | 0.051 | 0.3351 | No | ||
| 65 | SP4 | 8076 | -0.012 | 0.3214 | No | ||
| 66 | PDGFRA | 8362 | -0.075 | 0.3065 | No | ||
| 67 | TMEM47 | 8577 | -0.121 | 0.2958 | No | ||
| 68 | ISL1 | 8824 | -0.168 | 0.2836 | No | ||
| 69 | SGPL1 | 9042 | -0.217 | 0.2734 | No | ||
| 70 | ITSN1 | 9194 | -0.250 | 0.2670 | No | ||
| 71 | DCX | 9370 | -0.290 | 0.2595 | No | ||
| 72 | SERTAD1 | 9395 | -0.294 | 0.2603 | No | ||
| 73 | NUP35 | 9530 | -0.325 | 0.2553 | No | ||
| 74 | NFATC4 | 9593 | -0.339 | 0.2543 | No | ||
| 75 | ACY1L2 | 10012 | -0.416 | 0.2346 | No | ||
| 76 | NFIB | 10041 | -0.420 | 0.2360 | No | ||
| 77 | OSBPL3 | 10272 | -0.466 | 0.2268 | No | ||
| 78 | HOXA1 | 10385 | -0.487 | 0.2241 | No | ||
| 79 | SYNGR1 | 10893 | -0.582 | 0.2007 | No | ||
| 80 | FSTL1 | 11441 | -0.684 | 0.1759 | No | ||
| 81 | MEF2D | 11709 | -0.727 | 0.1665 | No | ||
| 82 | TNPO1 | 11858 | -0.754 | 0.1638 | No | ||
| 83 | SLC4A4 | 12624 | -0.894 | 0.1286 | No | ||
| 84 | KCND2 | 12797 | -0.924 | 0.1257 | No | ||
| 85 | ABCG4 | 12980 | -0.959 | 0.1225 | No | ||
| 86 | SNAP25 | 13123 | -0.983 | 0.1217 | No | ||
| 87 | MLLT6 | 13290 | -1.019 | 0.1198 | No | ||
| 88 | ETV1 | 13444 | -1.046 | 0.1187 | No | ||
| 89 | CPEB2 | 14049 | -1.163 | 0.0942 | No | ||
| 90 | SOX30 | 14387 | -1.239 | 0.0845 | No | ||
| 91 | FREQ | 14435 | -1.250 | 0.0907 | No | ||
| 92 | GDI1 | 14718 | -1.305 | 0.0845 | No | ||
| 93 | ETS1 | 14799 | -1.325 | 0.0893 | No | ||
| 94 | CLCF1 | 15095 | -1.393 | 0.0830 | No | ||
| 95 | ABCC3 | 15311 | -1.443 | 0.0814 | No | ||
| 96 | CSNK1G1 | 15464 | -1.482 | 0.0834 | No | ||
| 97 | APLN | 16449 | -1.764 | 0.0425 | No | ||
| 98 | TGFBR2 | 16674 | -1.846 | 0.0432 | No | ||
| 99 | EFNA3 | 17007 | -1.979 | 0.0389 | No | ||
| 100 | CLSTN2 | 17514 | -2.222 | 0.0270 | No | ||
| 101 | FGFR2 | 17588 | -2.265 | 0.0387 | No | ||
| 102 | DGCR2 | 17798 | -2.423 | 0.0442 | No |
