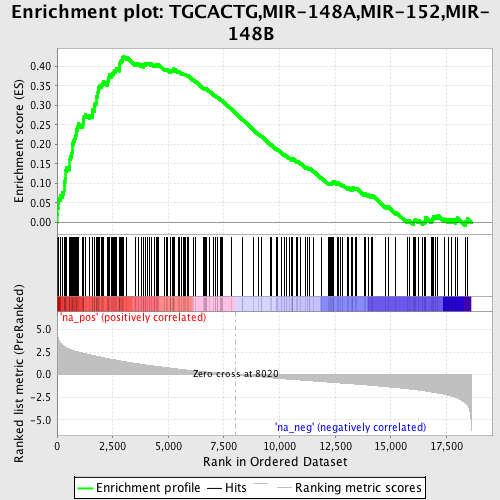
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TGCACTG,MIR-148A,MIR-152,MIR-148B |
| Enrichment Score (ES) | 0.4244869 |
| Normalized Enrichment Score (NES) | 2.1767457 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.8330458E-4 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARHGAP21 | 2 | 5.846 | 0.0202 | Yes | ||
| 2 | PIK3R3 | 12 | 4.890 | 0.0368 | Yes | ||
| 3 | TGFA | 49 | 4.065 | 0.0489 | Yes | ||
| 4 | POU4F2 | 82 | 3.788 | 0.0604 | Yes | ||
| 5 | ATP2A2 | 151 | 3.491 | 0.0688 | Yes | ||
| 6 | XPO4 | 233 | 3.265 | 0.0758 | Yes | ||
| 7 | SKP1A | 309 | 3.141 | 0.0826 | Yes | ||
| 8 | DNMT1 | 313 | 3.135 | 0.0934 | Yes | ||
| 9 | MOSPD1 | 318 | 3.130 | 0.1041 | Yes | ||
| 10 | ARPP-19 | 355 | 3.075 | 0.1128 | Yes | ||
| 11 | NARG1 | 373 | 3.040 | 0.1224 | Yes | ||
| 12 | LBR | 375 | 3.037 | 0.1330 | Yes | ||
| 13 | RNF12 | 440 | 2.944 | 0.1397 | Yes | ||
| 14 | CYB5R4 | 543 | 2.832 | 0.1440 | Yes | ||
| 15 | C1GALT1 | 572 | 2.805 | 0.1523 | Yes | ||
| 16 | ITSN2 | 574 | 2.802 | 0.1620 | Yes | ||
| 17 | MTMR12 | 609 | 2.771 | 0.1698 | Yes | ||
| 18 | RNF44 | 660 | 2.727 | 0.1765 | Yes | ||
| 19 | PLAA | 673 | 2.715 | 0.1853 | Yes | ||
| 20 | SFRS2IP | 680 | 2.710 | 0.1944 | Yes | ||
| 21 | UBR1 | 687 | 2.703 | 0.2035 | Yes | ||
| 22 | NLK | 741 | 2.665 | 0.2099 | Yes | ||
| 23 | ZFYVE26 | 802 | 2.612 | 0.2157 | Yes | ||
| 24 | PDIA3 | 818 | 2.602 | 0.2240 | Yes | ||
| 25 | ZBED4 | 857 | 2.576 | 0.2309 | Yes | ||
| 26 | OSBPL11 | 876 | 2.567 | 0.2388 | Yes | ||
| 27 | ARFIP1 | 920 | 2.534 | 0.2453 | Yes | ||
| 28 | PPP1CB | 945 | 2.520 | 0.2528 | Yes | ||
| 29 | CNOT4 | 1143 | 2.390 | 0.2504 | Yes | ||
| 30 | RALBP1 | 1165 | 2.381 | 0.2575 | Yes | ||
| 31 | ABCB7 | 1186 | 2.367 | 0.2647 | Yes | ||
| 32 | ST8SIA3 | 1207 | 2.354 | 0.2718 | Yes | ||
| 33 | RAB35 | 1261 | 2.324 | 0.2770 | Yes | ||
| 34 | YWHAB | 1472 | 2.202 | 0.2732 | Yes | ||
| 35 | ARL8B | 1576 | 2.142 | 0.2751 | Yes | ||
| 36 | ELF5 | 1594 | 2.134 | 0.2816 | Yes | ||
| 37 | PPP1R12A | 1602 | 2.128 | 0.2886 | Yes | ||
| 38 | RPS6KA5 | 1673 | 2.089 | 0.2921 | Yes | ||
| 39 | WNT1 | 1675 | 2.088 | 0.2993 | Yes | ||
| 40 | CDC14A | 1701 | 2.072 | 0.3051 | Yes | ||
| 41 | TOMM70A | 1770 | 2.039 | 0.3085 | Yes | ||
| 42 | PAPD4 | 1774 | 2.038 | 0.3155 | Yes | ||
| 43 | NCOA1 | 1776 | 2.036 | 0.3225 | Yes | ||
| 44 | LTBP1 | 1832 | 2.009 | 0.3265 | Yes | ||
| 45 | CFL2 | 1833 | 2.009 | 0.3335 | Yes | ||
| 46 | MLLT10 | 1842 | 2.004 | 0.3400 | Yes | ||
| 47 | TNRC6A | 1873 | 1.984 | 0.3453 | Yes | ||
| 48 | WDR20 | 1918 | 1.958 | 0.3497 | Yes | ||
| 49 | OTX1 | 1994 | 1.922 | 0.3523 | Yes | ||
| 50 | USP48 | 2043 | 1.893 | 0.3563 | Yes | ||
| 51 | GMFB | 2077 | 1.876 | 0.3610 | Yes | ||
| 52 | USP33 | 2265 | 1.789 | 0.3571 | Yes | ||
| 53 | BMP1 | 2283 | 1.779 | 0.3624 | Yes | ||
| 54 | HAS3 | 2309 | 1.766 | 0.3672 | Yes | ||
| 55 | DDX6 | 2326 | 1.756 | 0.3724 | Yes | ||
| 56 | MAX | 2333 | 1.753 | 0.3782 | Yes | ||
| 57 | MMD | 2465 | 1.693 | 0.3770 | Yes | ||
| 58 | ARRDC3 | 2496 | 1.680 | 0.3812 | Yes | ||
| 59 | BCL7A | 2544 | 1.659 | 0.3844 | Yes | ||
| 60 | KLF4 | 2558 | 1.654 | 0.3894 | Yes | ||
| 61 | CNTN4 | 2640 | 1.613 | 0.3906 | Yes | ||
| 62 | EIF2C1 | 2674 | 1.597 | 0.3944 | Yes | ||
| 63 | PDK4 | 2792 | 1.544 | 0.3934 | Yes | ||
| 64 | QKI | 2799 | 1.540 | 0.3985 | Yes | ||
| 65 | CPD | 2801 | 1.539 | 0.4038 | Yes | ||
| 66 | ARFGEF1 | 2817 | 1.532 | 0.4083 | Yes | ||
| 67 | PICALM | 2847 | 1.519 | 0.4120 | Yes | ||
| 68 | CABP7 | 2886 | 1.503 | 0.4151 | Yes | ||
| 69 | SMS | 2920 | 1.489 | 0.4185 | Yes | ||
| 70 | LMTK2 | 2948 | 1.476 | 0.4222 | Yes | ||
| 71 | SULF1 | 3000 | 1.452 | 0.4245 | Yes | ||
| 72 | TRAK2 | 3119 | 1.405 | 0.4230 | No | ||
| 73 | ARHGEF17 | 3504 | 1.254 | 0.4065 | No | ||
| 74 | CHD9 | 3543 | 1.235 | 0.4087 | No | ||
| 75 | EPN2 | 3652 | 1.193 | 0.4070 | No | ||
| 76 | B4GALT2 | 3779 | 1.144 | 0.4041 | No | ||
| 77 | NME7 | 3889 | 1.107 | 0.4021 | No | ||
| 78 | RAB34 | 3902 | 1.103 | 0.4052 | No | ||
| 79 | MEOX2 | 3961 | 1.079 | 0.4058 | No | ||
| 80 | RUNX2 | 3986 | 1.071 | 0.4083 | No | ||
| 81 | MAMDC1 | 4053 | 1.044 | 0.4083 | No | ||
| 82 | KLC2 | 4139 | 1.020 | 0.4072 | No | ||
| 83 | CDK5R1 | 4222 | 0.995 | 0.4063 | No | ||
| 84 | SIRT7 | 4355 | 0.951 | 0.4024 | No | ||
| 85 | SLC7A11 | 4396 | 0.939 | 0.4035 | No | ||
| 86 | GAP43 | 4446 | 0.923 | 0.4040 | No | ||
| 87 | CAMK2A | 4505 | 0.907 | 0.4040 | No | ||
| 88 | PPARGC1A | 4538 | 0.893 | 0.4054 | No | ||
| 89 | STARD13 | 4815 | 0.811 | 0.3932 | No | ||
| 90 | EIF2C4 | 4897 | 0.789 | 0.3916 | No | ||
| 91 | AKAP1 | 4951 | 0.770 | 0.3914 | No | ||
| 92 | INHBB | 5097 | 0.724 | 0.3860 | No | ||
| 93 | COL2A1 | 5100 | 0.723 | 0.3884 | No | ||
| 94 | DICER1 | 5111 | 0.720 | 0.3904 | No | ||
| 95 | RXRA | 5196 | 0.696 | 0.3883 | No | ||
| 96 | UCP3 | 5216 | 0.691 | 0.3896 | No | ||
| 97 | FOSB | 5227 | 0.687 | 0.3915 | No | ||
| 98 | MITF | 5235 | 0.686 | 0.3935 | No | ||
| 99 | FBXO33 | 5296 | 0.667 | 0.3925 | No | ||
| 100 | CLOCK | 5449 | 0.620 | 0.3864 | No | ||
| 101 | WNT10B | 5500 | 0.607 | 0.3858 | No | ||
| 102 | SIX4 | 5611 | 0.576 | 0.3819 | No | ||
| 103 | E2F3 | 5681 | 0.554 | 0.3801 | No | ||
| 104 | SLC25A3 | 5712 | 0.546 | 0.3803 | No | ||
| 105 | PSCD3 | 5756 | 0.536 | 0.3799 | No | ||
| 106 | BAZ2A | 5872 | 0.508 | 0.3754 | No | ||
| 107 | EFNB2 | 5884 | 0.503 | 0.3765 | No | ||
| 108 | E2F7 | 6120 | 0.447 | 0.3653 | No | ||
| 109 | RSBN1L | 6227 | 0.415 | 0.3610 | No | ||
| 110 | LIN28 | 6572 | 0.335 | 0.3435 | No | ||
| 111 | ZNRF1 | 6603 | 0.326 | 0.3430 | No | ||
| 112 | SSR1 | 6608 | 0.325 | 0.3439 | No | ||
| 113 | NOG | 6679 | 0.308 | 0.3412 | No | ||
| 114 | BRPF1 | 6686 | 0.306 | 0.3419 | No | ||
| 115 | MGAT4A | 6694 | 0.304 | 0.3426 | No | ||
| 116 | ATP7A | 6718 | 0.299 | 0.3424 | No | ||
| 117 | DNER | 6846 | 0.268 | 0.3364 | No | ||
| 118 | ADAMTS18 | 7032 | 0.226 | 0.3272 | No | ||
| 119 | TAF4 | 7105 | 0.211 | 0.3240 | No | ||
| 120 | PBXIP1 | 7111 | 0.210 | 0.3244 | No | ||
| 121 | UBE2D1 | 7208 | 0.187 | 0.3199 | No | ||
| 122 | SOX11 | 7214 | 0.185 | 0.3203 | No | ||
| 123 | ITGA5 | 7341 | 0.154 | 0.3139 | No | ||
| 124 | AKAP7 | 7352 | 0.152 | 0.3139 | No | ||
| 125 | MAFB | 7391 | 0.145 | 0.3124 | No | ||
| 126 | GPRC5B | 7392 | 0.144 | 0.3129 | No | ||
| 127 | CSF1 | 7439 | 0.135 | 0.3108 | No | ||
| 128 | MTF1 | 7448 | 0.133 | 0.3109 | No | ||
| 129 | PRICKLE2 | 7816 | 0.051 | 0.2911 | No | ||
| 130 | EDG1 | 8326 | -0.068 | 0.2637 | No | ||
| 131 | ZCCHC2 | 8334 | -0.069 | 0.2636 | No | ||
| 132 | ESRRG | 8806 | -0.163 | 0.2386 | No | ||
| 133 | ESR1 | 9054 | -0.220 | 0.2259 | No | ||
| 134 | RGMA | 9173 | -0.245 | 0.2204 | No | ||
| 135 | BCL11A | 9198 | -0.251 | 0.2199 | No | ||
| 136 | TGIF2 | 9608 | -0.342 | 0.1989 | No | ||
| 137 | BZRAP1 | 9625 | -0.346 | 0.1992 | No | ||
| 138 | PLEKHH1 | 9853 | -0.387 | 0.1883 | No | ||
| 139 | HMGB3 | 9923 | -0.400 | 0.1859 | No | ||
| 140 | JPH3 | 10095 | -0.432 | 0.1781 | No | ||
| 141 | FBN1 | 10201 | -0.453 | 0.1740 | No | ||
| 142 | ARHGEF12 | 10293 | -0.469 | 0.1707 | No | ||
| 143 | NPTX1 | 10441 | -0.496 | 0.1644 | No | ||
| 144 | EPS15 | 10542 | -0.516 | 0.1608 | No | ||
| 145 | RORB | 10560 | -0.520 | 0.1617 | No | ||
| 146 | NDST1 | 10587 | -0.525 | 0.1621 | No | ||
| 147 | TEAD1 | 10594 | -0.526 | 0.1636 | No | ||
| 148 | SNF1LK | 10775 | -0.559 | 0.1558 | No | ||
| 149 | MAF1 | 10814 | -0.567 | 0.1557 | No | ||
| 150 | TRPS1 | 10949 | -0.593 | 0.1505 | No | ||
| 151 | SYT1 | 11172 | -0.637 | 0.1406 | No | ||
| 152 | ACVR1 | 11232 | -0.648 | 0.1397 | No | ||
| 153 | LDLR | 11331 | -0.666 | 0.1367 | No | ||
| 154 | MAP3K9 | 11355 | -0.670 | 0.1377 | No | ||
| 155 | SLC24A3 | 11518 | -0.699 | 0.1314 | No | ||
| 156 | TMSB10 | 11887 | -0.758 | 0.1140 | No | ||
| 157 | MUM1L1 | 12188 | -0.814 | 0.1006 | No | ||
| 158 | PRKAA1 | 12247 | -0.826 | 0.1003 | No | ||
| 159 | ABCA1 | 12299 | -0.834 | 0.1004 | No | ||
| 160 | GPM6A | 12345 | -0.842 | 0.1009 | No | ||
| 161 | BTBD11 | 12377 | -0.848 | 0.1022 | No | ||
| 162 | BACH2 | 12401 | -0.851 | 0.1039 | No | ||
| 163 | ROBO2 | 12437 | -0.859 | 0.1050 | No | ||
| 164 | ANK2 | 12595 | -0.888 | 0.0995 | No | ||
| 165 | GADD45A | 12625 | -0.894 | 0.1011 | No | ||
| 166 | HOXC8 | 12751 | -0.918 | 0.0975 | No | ||
| 167 | ZFPM2 | 12845 | -0.934 | 0.0957 | No | ||
| 168 | GDF6 | 13058 | -0.973 | 0.0875 | No | ||
| 169 | CUL5 | 13081 | -0.976 | 0.0897 | No | ||
| 170 | ROBO1 | 13245 | -1.008 | 0.0844 | No | ||
| 171 | CHRD | 13289 | -1.019 | 0.0856 | No | ||
| 172 | MLLT6 | 13290 | -1.019 | 0.0891 | No | ||
| 173 | RNF38 | 13398 | -1.039 | 0.0869 | No | ||
| 174 | BTBD3 | 13471 | -1.053 | 0.0867 | No | ||
| 175 | B4GALT5 | 13811 | -1.115 | 0.0722 | No | ||
| 176 | ERBB3 | 13857 | -1.124 | 0.0736 | No | ||
| 177 | DPP3 | 13995 | -1.151 | 0.0702 | No | ||
| 178 | DYRK1B | 14123 | -1.179 | 0.0674 | No | ||
| 179 | ITPK1 | 14177 | -1.191 | 0.0687 | No | ||
| 180 | PTPRM | 14768 | -1.319 | 0.0412 | No | ||
| 181 | SEPT4 | 14875 | -1.342 | 0.0401 | No | ||
| 182 | PHACTR2 | 15229 | -1.422 | 0.0259 | No | ||
| 183 | ATXN1 | 15736 | -1.552 | 0.0038 | No | ||
| 184 | TRPM2 | 15836 | -1.583 | 0.0039 | No | ||
| 185 | CHD7 | 16016 | -1.635 | -0.0001 | No | ||
| 186 | CANX | 16041 | -1.642 | 0.0043 | No | ||
| 187 | PCDH10 | 16089 | -1.656 | 0.0075 | No | ||
| 188 | DEDD | 16250 | -1.696 | 0.0047 | No | ||
| 189 | FBXL19 | 16445 | -1.762 | 0.0003 | No | ||
| 190 | YPEL3 | 16508 | -1.785 | 0.0032 | No | ||
| 191 | SLC2A1 | 16557 | -1.804 | 0.0068 | No | ||
| 192 | POU3F2 | 16562 | -1.806 | 0.0129 | No | ||
| 193 | PRKAG2 | 16831 | -1.912 | 0.0050 | No | ||
| 194 | FMR1 | 16887 | -1.931 | 0.0087 | No | ||
| 195 | MMP19 | 16913 | -1.943 | 0.0141 | No | ||
| 196 | ITGA11 | 17009 | -1.980 | 0.0159 | No | ||
| 197 | KLHL18 | 17115 | -2.026 | 0.0172 | No | ||
| 198 | ALS2CR2 | 17415 | -2.166 | 0.0085 | No | ||
| 199 | ADCY2 | 17595 | -2.269 | 0.0067 | No | ||
| 200 | SNPH | 17737 | -2.371 | 0.0073 | No | ||
| 201 | ELMO1 | 17910 | -2.505 | 0.0067 | No | ||
| 202 | MNT | 17992 | -2.589 | 0.0113 | No | ||
| 203 | HRB | 18373 | -3.167 | 0.0016 | No | ||
| 204 | ETV5 | 18434 | -3.305 | 0.0099 | No |
