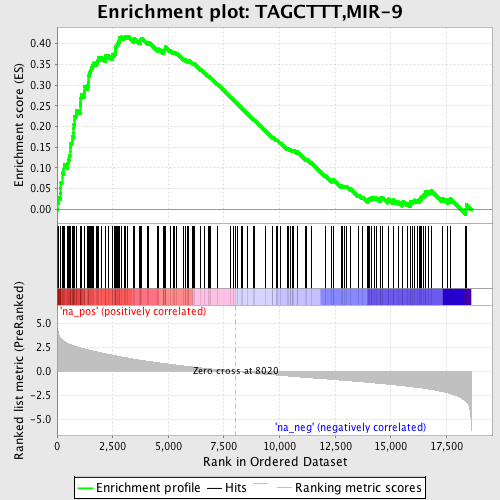
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TAGCTTT,MIR-9 |
| Enrichment Score (ES) | 0.4182859 |
| Normalized Enrichment Score (NES) | 2.0801263 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.312011E-4 |
| FWER p-Value | 0.011 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ERBB2IP | 29 | 4.297 | 0.0154 | Yes | ||
| 2 | PCAF | 66 | 3.924 | 0.0290 | Yes | ||
| 3 | PPP3R1 | 143 | 3.514 | 0.0388 | Yes | ||
| 4 | MAGI1 | 158 | 3.456 | 0.0517 | Yes | ||
| 5 | ELOVL6 | 171 | 3.425 | 0.0647 | Yes | ||
| 6 | MECP2 | 220 | 3.287 | 0.0751 | Yes | ||
| 7 | HNRPA2B1 | 221 | 3.287 | 0.0881 | Yes | ||
| 8 | TSGA14 | 302 | 3.153 | 0.0962 | Yes | ||
| 9 | SKP1A | 309 | 3.141 | 0.1083 | Yes | ||
| 10 | SLITRK3 | 481 | 2.904 | 0.1106 | Yes | ||
| 11 | KPNB1 | 493 | 2.884 | 0.1214 | Yes | ||
| 12 | FRAS1 | 544 | 2.831 | 0.1299 | Yes | ||
| 13 | ZHX1 | 581 | 2.796 | 0.1390 | Yes | ||
| 14 | FYCO1 | 584 | 2.794 | 0.1499 | Yes | ||
| 15 | MTMR12 | 609 | 2.771 | 0.1596 | Yes | ||
| 16 | NDEL1 | 670 | 2.716 | 0.1671 | Yes | ||
| 17 | CUGBP2 | 695 | 2.698 | 0.1765 | Yes | ||
| 18 | NLK | 741 | 2.665 | 0.1846 | Yes | ||
| 19 | RAB14 | 746 | 2.665 | 0.1949 | Yes | ||
| 20 | KRIT1 | 755 | 2.657 | 0.2050 | Yes | ||
| 21 | ANKRD10 | 785 | 2.625 | 0.2139 | Yes | ||
| 22 | SFMBT1 | 788 | 2.622 | 0.2241 | Yes | ||
| 23 | STRN3 | 851 | 2.578 | 0.2310 | Yes | ||
| 24 | SLC25A37 | 885 | 2.561 | 0.2393 | Yes | ||
| 25 | MIB1 | 1033 | 2.459 | 0.2411 | Yes | ||
| 26 | TRIM33 | 1034 | 2.459 | 0.2508 | Yes | ||
| 27 | STK4 | 1059 | 2.443 | 0.2592 | Yes | ||
| 28 | SFRS2 | 1068 | 2.439 | 0.2684 | Yes | ||
| 29 | PSMD7 | 1087 | 2.431 | 0.2771 | Yes | ||
| 30 | KRAS | 1213 | 2.349 | 0.2796 | Yes | ||
| 31 | EIF4ENIF1 | 1230 | 2.341 | 0.2880 | Yes | ||
| 32 | CYR61 | 1235 | 2.338 | 0.2970 | Yes | ||
| 33 | RBBP5 | 1374 | 2.254 | 0.2985 | Yes | ||
| 34 | ETF1 | 1405 | 2.239 | 0.3057 | Yes | ||
| 35 | RAP2C | 1416 | 2.232 | 0.3140 | Yes | ||
| 36 | KCTD9 | 1417 | 2.230 | 0.3228 | Yes | ||
| 37 | MAP3K7 | 1445 | 2.215 | 0.3301 | Yes | ||
| 38 | APRIN | 1502 | 2.181 | 0.3357 | Yes | ||
| 39 | UBE2D3 | 1526 | 2.167 | 0.3431 | Yes | ||
| 40 | VAMP4 | 1601 | 2.129 | 0.3475 | Yes | ||
| 41 | PHF6 | 1646 | 2.099 | 0.3534 | Yes | ||
| 42 | HIP2 | 1753 | 2.053 | 0.3558 | Yes | ||
| 43 | CFL2 | 1833 | 2.009 | 0.3595 | Yes | ||
| 44 | BCL11B | 1857 | 1.998 | 0.3661 | Yes | ||
| 45 | SENP1 | 1974 | 1.931 | 0.3675 | Yes | ||
| 46 | ACTL6A | 2163 | 1.832 | 0.3645 | Yes | ||
| 47 | YWHAZ | 2185 | 1.824 | 0.3706 | Yes | ||
| 48 | SAR1B | 2291 | 1.775 | 0.3720 | Yes | ||
| 49 | LRRC4 | 2485 | 1.684 | 0.3682 | Yes | ||
| 50 | DDX3X | 2492 | 1.681 | 0.3745 | Yes | ||
| 51 | RARG | 2584 | 1.643 | 0.3761 | Yes | ||
| 52 | GSPT1 | 2613 | 1.629 | 0.3810 | Yes | ||
| 53 | SOX4 | 2626 | 1.618 | 0.3868 | Yes | ||
| 54 | CNTN4 | 2640 | 1.613 | 0.3924 | Yes | ||
| 55 | ZMYND11 | 2677 | 1.594 | 0.3968 | Yes | ||
| 56 | HMGB2 | 2708 | 1.582 | 0.4014 | Yes | ||
| 57 | ATP11A | 2767 | 1.557 | 0.4045 | Yes | ||
| 58 | QKI | 2799 | 1.540 | 0.4089 | Yes | ||
| 59 | TCEA1 | 2810 | 1.535 | 0.4144 | Yes | ||
| 60 | CYGB | 2885 | 1.503 | 0.4163 | Yes | ||
| 61 | PHF2 | 3006 | 1.449 | 0.4156 | Yes | ||
| 62 | ARNTL | 3076 | 1.419 | 0.4175 | Yes | ||
| 63 | YAF2 | 3163 | 1.388 | 0.4183 | Yes | ||
| 64 | NR3C1 | 3444 | 1.276 | 0.4082 | No | ||
| 65 | CASK | 3468 | 1.265 | 0.4119 | No | ||
| 66 | HNRPH2 | 3700 | 1.174 | 0.4040 | No | ||
| 67 | PDK1 | 3757 | 1.155 | 0.4056 | No | ||
| 68 | CIT | 3762 | 1.153 | 0.4099 | No | ||
| 69 | TIMP3 | 3801 | 1.139 | 0.4124 | No | ||
| 70 | NEGR1 | 4084 | 1.035 | 0.4012 | No | ||
| 71 | SORCS3 | 4121 | 1.023 | 0.4033 | No | ||
| 72 | THRB | 4514 | 0.903 | 0.3856 | No | ||
| 73 | STC1 | 4549 | 0.889 | 0.3873 | No | ||
| 74 | NR4A3 | 4766 | 0.823 | 0.3788 | No | ||
| 75 | PCTK2 | 4792 | 0.817 | 0.3807 | No | ||
| 76 | KCNJ3 | 4795 | 0.816 | 0.3838 | No | ||
| 77 | EPHA7 | 4832 | 0.807 | 0.3851 | No | ||
| 78 | HS6ST2 | 4849 | 0.803 | 0.3874 | No | ||
| 79 | SORCS1 | 4859 | 0.801 | 0.3901 | No | ||
| 80 | ZFP36L1 | 4874 | 0.797 | 0.3925 | No | ||
| 81 | AKAP2 | 5074 | 0.730 | 0.3846 | No | ||
| 82 | MITF | 5235 | 0.686 | 0.3786 | No | ||
| 83 | FBXO33 | 5296 | 0.667 | 0.3780 | No | ||
| 84 | ZIC2 | 5362 | 0.643 | 0.3770 | No | ||
| 85 | E2F3 | 5681 | 0.554 | 0.3620 | No | ||
| 86 | PSCD3 | 5756 | 0.536 | 0.3601 | No | ||
| 87 | LRP4 | 5762 | 0.534 | 0.3619 | No | ||
| 88 | DR1 | 5882 | 0.503 | 0.3575 | No | ||
| 89 | TARDBP | 5888 | 0.503 | 0.3592 | No | ||
| 90 | GOLGA4 | 5903 | 0.499 | 0.3604 | No | ||
| 91 | SLITRK2 | 6074 | 0.459 | 0.3530 | No | ||
| 92 | E2F7 | 6120 | 0.447 | 0.3523 | No | ||
| 93 | GABRA1 | 6162 | 0.434 | 0.3518 | No | ||
| 94 | HIPK1 | 6457 | 0.362 | 0.3373 | No | ||
| 95 | INSM1 | 6628 | 0.322 | 0.3294 | No | ||
| 96 | NFKBIZ | 6807 | 0.277 | 0.3208 | No | ||
| 97 | PCDH18 | 6839 | 0.270 | 0.3202 | No | ||
| 98 | MMP16 | 6898 | 0.255 | 0.3181 | No | ||
| 99 | DVL2 | 7209 | 0.187 | 0.3020 | No | ||
| 100 | TLL2 | 7787 | 0.057 | 0.2709 | No | ||
| 101 | CCND2 | 7931 | 0.024 | 0.2633 | No | ||
| 102 | SOCS4 | 8034 | -0.002 | 0.2578 | No | ||
| 103 | SP5 | 8110 | -0.019 | 0.2538 | No | ||
| 104 | SMARCA1 | 8272 | -0.053 | 0.2453 | No | ||
| 105 | EDG1 | 8326 | -0.068 | 0.2426 | No | ||
| 106 | TMEM47 | 8577 | -0.121 | 0.2296 | No | ||
| 107 | ZFHX4 | 8805 | -0.163 | 0.2179 | No | ||
| 108 | PIGF | 8871 | -0.180 | 0.2151 | No | ||
| 109 | DCX | 9370 | -0.290 | 0.1892 | No | ||
| 110 | ARHGAP5 | 9685 | -0.356 | 0.1736 | No | ||
| 111 | DPYSL3 | 9694 | -0.358 | 0.1746 | No | ||
| 112 | PARD6B | 9867 | -0.390 | 0.1668 | No | ||
| 113 | CAMK2G | 9889 | -0.394 | 0.1673 | No | ||
| 114 | MEF2C | 10048 | -0.422 | 0.1604 | No | ||
| 115 | PPP1R8 | 10333 | -0.477 | 0.1468 | No | ||
| 116 | SLC16A1 | 10381 | -0.486 | 0.1462 | No | ||
| 117 | SLC9A9 | 10478 | -0.504 | 0.1430 | No | ||
| 118 | NAP1L5 | 10499 | -0.508 | 0.1439 | No | ||
| 119 | NDST1 | 10587 | -0.525 | 0.1413 | No | ||
| 120 | TEAD1 | 10594 | -0.526 | 0.1431 | No | ||
| 121 | LRP12 | 10632 | -0.534 | 0.1432 | No | ||
| 122 | SET | 10787 | -0.561 | 0.1370 | No | ||
| 123 | STAT1 | 10791 | -0.562 | 0.1391 | No | ||
| 124 | AKT3 | 11164 | -0.635 | 0.1214 | No | ||
| 125 | GABRB3 | 11208 | -0.644 | 0.1217 | No | ||
| 126 | CRISPLD1 | 11426 | -0.682 | 0.1126 | No | ||
| 127 | PRKCE | 12056 | -0.791 | 0.0816 | No | ||
| 128 | GPM6A | 12345 | -0.842 | 0.0693 | No | ||
| 129 | BACH2 | 12401 | -0.851 | 0.0697 | No | ||
| 130 | PRPF4B | 12408 | -0.852 | 0.0728 | No | ||
| 131 | TPD52 | 12761 | -0.919 | 0.0573 | No | ||
| 132 | ZFPM2 | 12845 | -0.934 | 0.0565 | No | ||
| 133 | SOX3 | 12930 | -0.950 | 0.0557 | No | ||
| 134 | OSBPL8 | 12990 | -0.960 | 0.0563 | No | ||
| 135 | ENAH | 13181 | -0.997 | 0.0500 | No | ||
| 136 | PITPNM2 | 13542 | -1.068 | 0.0347 | No | ||
| 137 | HOXB5 | 13718 | -1.098 | 0.0296 | No | ||
| 138 | STX16 | 13947 | -1.142 | 0.0217 | No | ||
| 139 | GSPT2 | 13984 | -1.149 | 0.0243 | No | ||
| 140 | CPEB2 | 14049 | -1.163 | 0.0254 | No | ||
| 141 | SERF2 | 14126 | -1.179 | 0.0260 | No | ||
| 142 | ELAVL2 | 14146 | -1.184 | 0.0297 | No | ||
| 143 | LRRTM1 | 14266 | -1.212 | 0.0280 | No | ||
| 144 | FBXO22 | 14361 | -1.235 | 0.0278 | No | ||
| 145 | SOX2 | 14529 | -1.272 | 0.0238 | No | ||
| 146 | NCKIPSD | 14549 | -1.275 | 0.0278 | No | ||
| 147 | HOXB13 | 14620 | -1.289 | 0.0291 | No | ||
| 148 | TPM1 | 14873 | -1.341 | 0.0207 | No | ||
| 149 | DNAJC6 | 14889 | -1.345 | 0.0253 | No | ||
| 150 | CPEB3 | 15108 | -1.397 | 0.0190 | No | ||
| 151 | NR2F2 | 15134 | -1.401 | 0.0232 | No | ||
| 152 | AFF3 | 15335 | -1.447 | 0.0180 | No | ||
| 153 | WAC | 15519 | -1.498 | 0.0141 | No | ||
| 154 | SOX21 | 15545 | -1.503 | 0.0186 | No | ||
| 155 | ATXN1 | 15736 | -1.552 | 0.0145 | No | ||
| 156 | PI15 | 15871 | -1.592 | 0.0135 | No | ||
| 157 | PLCG1 | 15887 | -1.596 | 0.0190 | No | ||
| 158 | MPP5 | 15985 | -1.627 | 0.0202 | No | ||
| 159 | THBD | 16059 | -1.646 | 0.0228 | No | ||
| 160 | CALD1 | 16193 | -1.681 | 0.0222 | No | ||
| 161 | POU4F1 | 16280 | -1.704 | 0.0243 | No | ||
| 162 | CREB5 | 16342 | -1.724 | 0.0278 | No | ||
| 163 | CRK | 16397 | -1.743 | 0.0318 | No | ||
| 164 | ITGB1 | 16469 | -1.768 | 0.0349 | No | ||
| 165 | KCNN2 | 16556 | -1.804 | 0.0374 | No | ||
| 166 | POU3F2 | 16562 | -1.806 | 0.0443 | No | ||
| 167 | DTX3 | 16711 | -1.859 | 0.0436 | No | ||
| 168 | GPHN | 16814 | -1.905 | 0.0457 | No | ||
| 169 | PHF16 | 17325 | -2.116 | 0.0264 | No | ||
| 170 | HTR2C | 17552 | -2.244 | 0.0230 | No | ||
| 171 | SLITRK6 | 17668 | -2.320 | 0.0260 | No | ||
| 172 | HRB | 18373 | -3.167 | 0.0003 | No | ||
| 173 | TNFRSF19L | 18403 | -3.231 | 0.0115 | No |
