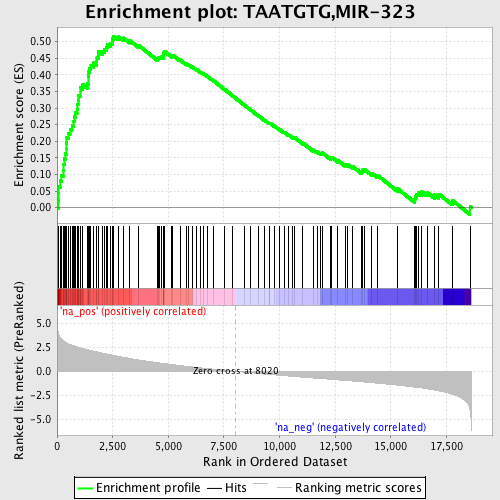
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | 0.5165587 |
| Normalized Enrichment Score (NES) | 2.3894308 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TGFA | 49 | 4.065 | 0.0212 | Yes | ||
| 2 | CCND1 | 71 | 3.865 | 0.0427 | Yes | ||
| 3 | POU4F2 | 82 | 3.788 | 0.0643 | Yes | ||
| 4 | ATP11B | 133 | 3.537 | 0.0823 | Yes | ||
| 5 | SATB1 | 202 | 3.343 | 0.0982 | Yes | ||
| 6 | SFRS1 | 283 | 3.183 | 0.1126 | Yes | ||
| 7 | REV3L | 304 | 3.147 | 0.1299 | Yes | ||
| 8 | SMARCAD1 | 326 | 3.118 | 0.1470 | Yes | ||
| 9 | PDCD10 | 377 | 3.036 | 0.1621 | Yes | ||
| 10 | TOP2B | 406 | 2.984 | 0.1781 | Yes | ||
| 11 | NFAT5 | 408 | 2.982 | 0.1955 | Yes | ||
| 12 | RAB1A | 439 | 2.944 | 0.2111 | Yes | ||
| 13 | TOP1 | 502 | 2.872 | 0.2246 | Yes | ||
| 14 | ZHX1 | 581 | 2.796 | 0.2368 | Yes | ||
| 15 | NDEL1 | 670 | 2.716 | 0.2479 | Yes | ||
| 16 | KPNA1 | 728 | 2.676 | 0.2605 | Yes | ||
| 17 | DHX36 | 768 | 2.642 | 0.2739 | Yes | ||
| 18 | RANBP9 | 833 | 2.593 | 0.2856 | Yes | ||
| 19 | DCUN1D1 | 897 | 2.551 | 0.2972 | Yes | ||
| 20 | ARFIP1 | 920 | 2.534 | 0.3108 | Yes | ||
| 21 | PPP1CB | 945 | 2.520 | 0.3243 | Yes | ||
| 22 | ZNF318 | 974 | 2.500 | 0.3374 | Yes | ||
| 23 | TRIM33 | 1034 | 2.459 | 0.3486 | Yes | ||
| 24 | HS2ST1 | 1036 | 2.459 | 0.3630 | Yes | ||
| 25 | SUMO1 | 1156 | 2.384 | 0.3705 | Yes | ||
| 26 | FEM1B | 1357 | 2.264 | 0.3730 | Yes | ||
| 27 | CEPT1 | 1394 | 2.244 | 0.3842 | Yes | ||
| 28 | PJA2 | 1395 | 2.243 | 0.3973 | Yes | ||
| 29 | ZDHHC21 | 1425 | 2.227 | 0.4088 | Yes | ||
| 30 | SLMAP | 1469 | 2.204 | 0.4194 | Yes | ||
| 31 | MAPRE1 | 1521 | 2.168 | 0.4293 | Yes | ||
| 32 | CLCN3 | 1631 | 2.113 | 0.4358 | Yes | ||
| 33 | CUL1 | 1780 | 2.035 | 0.4397 | Yes | ||
| 34 | UBL3 | 1791 | 2.031 | 0.4511 | Yes | ||
| 35 | UHRF2 | 1844 | 2.003 | 0.4600 | Yes | ||
| 36 | TNRC6A | 1873 | 1.984 | 0.4701 | Yes | ||
| 37 | CSNK1G3 | 2049 | 1.891 | 0.4717 | Yes | ||
| 38 | PITPNA | 2142 | 1.842 | 0.4776 | Yes | ||
| 39 | ASF1A | 2223 | 1.808 | 0.4838 | Yes | ||
| 40 | RSBN1 | 2256 | 1.791 | 0.4926 | Yes | ||
| 41 | HOOK3 | 2412 | 1.717 | 0.4943 | Yes | ||
| 42 | LRRC4 | 2485 | 1.684 | 0.5002 | Yes | ||
| 43 | DDX3X | 2492 | 1.681 | 0.5098 | Yes | ||
| 44 | ULK2 | 2547 | 1.657 | 0.5166 | Yes | ||
| 45 | RPS6KA3 | 2760 | 1.562 | 0.5143 | No | ||
| 46 | TMPO | 2980 | 1.460 | 0.5110 | No | ||
| 47 | STX12 | 3262 | 1.343 | 0.5037 | No | ||
| 48 | RAB10 | 3674 | 1.183 | 0.4884 | No | ||
| 49 | WTAP | 4510 | 0.905 | 0.4486 | No | ||
| 50 | PPARGC1A | 4538 | 0.893 | 0.4523 | No | ||
| 51 | ACAT2 | 4622 | 0.862 | 0.4529 | No | ||
| 52 | NFIX | 4696 | 0.840 | 0.4539 | No | ||
| 53 | NR4A3 | 4766 | 0.823 | 0.4550 | No | ||
| 54 | UTX | 4780 | 0.819 | 0.4591 | No | ||
| 55 | KCNJ3 | 4795 | 0.816 | 0.4631 | No | ||
| 56 | TBPL1 | 4801 | 0.815 | 0.4676 | No | ||
| 57 | MRC1 | 4841 | 0.805 | 0.4702 | No | ||
| 58 | CLASP1 | 5155 | 0.707 | 0.4574 | No | ||
| 59 | USP9X | 5192 | 0.697 | 0.4596 | No | ||
| 60 | AUH | 5526 | 0.600 | 0.4451 | No | ||
| 61 | EBF3 | 5804 | 0.523 | 0.4332 | No | ||
| 62 | NRK | 5913 | 0.496 | 0.4303 | No | ||
| 63 | SLITRK2 | 6074 | 0.459 | 0.4243 | No | ||
| 64 | KPNA3 | 6286 | 0.403 | 0.4153 | No | ||
| 65 | HIPK1 | 6457 | 0.362 | 0.4082 | No | ||
| 66 | RCN1 | 6591 | 0.331 | 0.4029 | No | ||
| 67 | CHCHD4 | 6755 | 0.290 | 0.3958 | No | ||
| 68 | MAP4K4 | 7030 | 0.226 | 0.3824 | No | ||
| 69 | ENTPD5 | 7544 | 0.110 | 0.3553 | No | ||
| 70 | WBP2 | 7874 | 0.037 | 0.3377 | No | ||
| 71 | GJA1 | 8400 | -0.082 | 0.3098 | No | ||
| 72 | ZIC1 | 8692 | -0.143 | 0.2950 | No | ||
| 73 | RAB11FIP2 | 9044 | -0.217 | 0.2773 | No | ||
| 74 | SPSB4 | 9338 | -0.284 | 0.2631 | No | ||
| 75 | OSBP | 9535 | -0.327 | 0.2544 | No | ||
| 76 | ANKRD27 | 9562 | -0.331 | 0.2550 | No | ||
| 77 | STK35 | 9766 | -0.373 | 0.2462 | No | ||
| 78 | PCDH9 | 10005 | -0.414 | 0.2357 | No | ||
| 79 | FBN1 | 10201 | -0.453 | 0.2279 | No | ||
| 80 | HOXA1 | 10385 | -0.487 | 0.2208 | No | ||
| 81 | TEAD1 | 10594 | -0.526 | 0.2127 | No | ||
| 82 | QSCN6L1 | 10690 | -0.543 | 0.2107 | No | ||
| 83 | NFKB1 | 11045 | -0.612 | 0.1952 | No | ||
| 84 | POU2F3 | 11516 | -0.699 | 0.1739 | No | ||
| 85 | LPP | 11687 | -0.723 | 0.1689 | No | ||
| 86 | TMOD1 | 11851 | -0.752 | 0.1645 | No | ||
| 87 | EIF3S1 | 11907 | -0.762 | 0.1660 | No | ||
| 88 | DPF2 | 12289 | -0.833 | 0.1503 | No | ||
| 89 | GPM6A | 12345 | -0.842 | 0.1523 | No | ||
| 90 | SLC4A4 | 12624 | -0.894 | 0.1425 | No | ||
| 91 | BET1 | 12945 | -0.953 | 0.1308 | No | ||
| 92 | CXCL12 | 13053 | -0.972 | 0.1307 | No | ||
| 93 | TMEM16D | 13276 | -1.015 | 0.1246 | No | ||
| 94 | STAT5B | 13671 | -1.090 | 0.1097 | No | ||
| 95 | HOXB5 | 13718 | -1.098 | 0.1137 | No | ||
| 96 | FGD4 | 13806 | -1.114 | 0.1155 | No | ||
| 97 | CDKN1B | 14151 | -1.185 | 0.1039 | No | ||
| 98 | MYLIP | 14395 | -1.242 | 0.0980 | No | ||
| 99 | MAP3K3 | 15290 | -1.438 | 0.0581 | No | ||
| 100 | SEPT3 | 16074 | -1.651 | 0.0255 | No | ||
| 101 | PKP4 | 16103 | -1.659 | 0.0337 | No | ||
| 102 | EGR3 | 16167 | -1.673 | 0.0401 | No | ||
| 103 | PAK7 | 16238 | -1.693 | 0.0462 | No | ||
| 104 | CRK | 16397 | -1.743 | 0.0479 | No | ||
| 105 | MCFD2 | 16631 | -1.829 | 0.0460 | No | ||
| 106 | SEMA6D | 16975 | -1.966 | 0.0390 | No | ||
| 107 | FBN2 | 17164 | -2.042 | 0.0408 | No | ||
| 108 | CREB1 | 17762 | -2.389 | 0.0225 | No | ||
| 109 | PTK2B | 18558 | -4.019 | 0.0031 | No |
