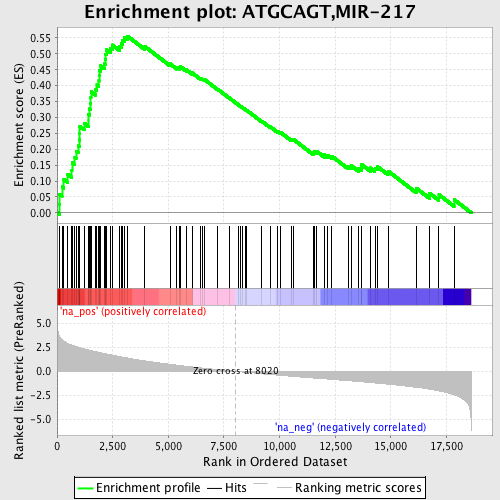
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ATGCAGT,MIR-217 |
| Enrichment Score (ES) | 0.5557098 |
| Normalized Enrichment Score (NES) | 2.4366326 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHMT1 | 98 | 3.704 | 0.0271 | Yes | ||
| 2 | APPBP2 | 106 | 3.674 | 0.0589 | Yes | ||
| 3 | HNRPA2B1 | 221 | 3.287 | 0.0814 | Yes | ||
| 4 | PPM1D | 305 | 3.147 | 0.1045 | Yes | ||
| 5 | SLITRK3 | 481 | 2.904 | 0.1204 | Yes | ||
| 6 | OGT | 665 | 2.718 | 0.1343 | Yes | ||
| 7 | NDEL1 | 670 | 2.716 | 0.1579 | Yes | ||
| 8 | TLK2 | 800 | 2.616 | 0.1738 | Yes | ||
| 9 | SLC31A1 | 853 | 2.577 | 0.1935 | Yes | ||
| 10 | SLC39A10 | 949 | 2.516 | 0.2104 | Yes | ||
| 11 | MYEF2 | 1005 | 2.483 | 0.2291 | Yes | ||
| 12 | MTPN | 1016 | 2.477 | 0.2502 | Yes | ||
| 13 | EIF4A2 | 1023 | 2.470 | 0.2715 | Yes | ||
| 14 | KRAS | 1213 | 2.349 | 0.2818 | Yes | ||
| 15 | RAP2C | 1416 | 2.232 | 0.2905 | Yes | ||
| 16 | KCTD9 | 1417 | 2.230 | 0.3100 | Yes | ||
| 17 | MAPK1 | 1466 | 2.206 | 0.3267 | Yes | ||
| 18 | APRIN | 1502 | 2.181 | 0.3438 | Yes | ||
| 19 | ZFYVE20 | 1517 | 2.170 | 0.3621 | Yes | ||
| 20 | PCNA | 1525 | 2.167 | 0.3806 | Yes | ||
| 21 | SIRT1 | 1739 | 2.056 | 0.3871 | Yes | ||
| 22 | UBL3 | 1791 | 2.031 | 0.4021 | Yes | ||
| 23 | ATP6V1A | 1880 | 1.981 | 0.4147 | Yes | ||
| 24 | YWHAG | 1884 | 1.980 | 0.4319 | Yes | ||
| 25 | MSN | 1893 | 1.970 | 0.4487 | Yes | ||
| 26 | HNRPUL1 | 1959 | 1.935 | 0.4621 | Yes | ||
| 27 | DYNC1LI2 | 2130 | 1.848 | 0.4691 | Yes | ||
| 28 | EZH2 | 2166 | 1.831 | 0.4832 | Yes | ||
| 29 | STAG2 | 2183 | 1.825 | 0.4983 | Yes | ||
| 30 | ATP1B1 | 2217 | 1.812 | 0.5123 | Yes | ||
| 31 | SLC38A2 | 2396 | 1.723 | 0.5178 | Yes | ||
| 32 | IVNS1ABP | 2484 | 1.685 | 0.5278 | Yes | ||
| 33 | ATP11C | 2797 | 1.541 | 0.5245 | Yes | ||
| 34 | RTF1 | 2891 | 1.499 | 0.5326 | Yes | ||
| 35 | SENP5 | 2951 | 1.475 | 0.5423 | Yes | ||
| 36 | HIVEP3 | 3026 | 1.443 | 0.5509 | Yes | ||
| 37 | YAF2 | 3163 | 1.388 | 0.5557 | Yes | ||
| 38 | ELOVL4 | 3947 | 1.086 | 0.5230 | No | ||
| 39 | AKAP2 | 5074 | 0.730 | 0.4686 | No | ||
| 40 | DYRK1A | 5380 | 0.637 | 0.4577 | No | ||
| 41 | STX1A | 5486 | 0.611 | 0.4574 | No | ||
| 42 | FN1 | 5535 | 0.597 | 0.4600 | No | ||
| 43 | JOSD3 | 5805 | 0.523 | 0.4501 | No | ||
| 44 | PSMF1 | 6064 | 0.462 | 0.4402 | No | ||
| 45 | DACH1 | 6447 | 0.364 | 0.4228 | No | ||
| 46 | GRIK2 | 6542 | 0.341 | 0.4207 | No | ||
| 47 | GDI2 | 6624 | 0.322 | 0.4192 | No | ||
| 48 | SOX11 | 7214 | 0.185 | 0.3890 | No | ||
| 49 | FBXW11 | 7737 | 0.066 | 0.3614 | No | ||
| 50 | RIN2 | 8165 | -0.031 | 0.3387 | No | ||
| 51 | ADAMTS5 | 8243 | -0.048 | 0.3349 | No | ||
| 52 | ZCCHC2 | 8334 | -0.069 | 0.3307 | No | ||
| 53 | DMRT2 | 8455 | -0.094 | 0.3251 | No | ||
| 54 | STX6 | 8510 | -0.108 | 0.3231 | No | ||
| 55 | BCL11A | 9198 | -0.251 | 0.2882 | No | ||
| 56 | MAMDC2 | 9587 | -0.338 | 0.2702 | No | ||
| 57 | NFIA | 9926 | -0.401 | 0.2555 | No | ||
| 58 | TMSB4X | 10020 | -0.417 | 0.2541 | No | ||
| 59 | ST18 | 10534 | -0.515 | 0.2310 | No | ||
| 60 | ACVR2A | 10621 | -0.531 | 0.2310 | No | ||
| 61 | BAI3 | 11517 | -0.699 | 0.1888 | No | ||
| 62 | PAQR3 | 11577 | -0.708 | 0.1918 | No | ||
| 63 | NEUROD1 | 11657 | -0.720 | 0.1938 | No | ||
| 64 | STRBP | 12002 | -0.782 | 0.1821 | No | ||
| 65 | DLGAP2 | 12143 | -0.805 | 0.1816 | No | ||
| 66 | GPM6A | 12345 | -0.842 | 0.1781 | No | ||
| 67 | CUL5 | 13081 | -0.976 | 0.1470 | No | ||
| 68 | A2BP1 | 13231 | -1.005 | 0.1478 | No | ||
| 69 | POLG | 13559 | -1.071 | 0.1395 | No | ||
| 70 | STAT5B | 13671 | -1.090 | 0.1430 | No | ||
| 71 | KCNA3 | 13684 | -1.092 | 0.1519 | No | ||
| 72 | CHN2 | 14076 | -1.167 | 0.1410 | No | ||
| 73 | NR4A2 | 14288 | -1.217 | 0.1403 | No | ||
| 74 | TBC1D15 | 14388 | -1.240 | 0.1458 | No | ||
| 75 | SEPT4 | 14875 | -1.342 | 0.1313 | No | ||
| 76 | CAPN3 | 16137 | -1.666 | 0.0779 | No | ||
| 77 | KLF12 | 16747 | -1.873 | 0.0614 | No | ||
| 78 | FBN2 | 17164 | -2.042 | 0.0568 | No | ||
| 79 | TACC2 | 17842 | -2.456 | 0.0418 | No |
