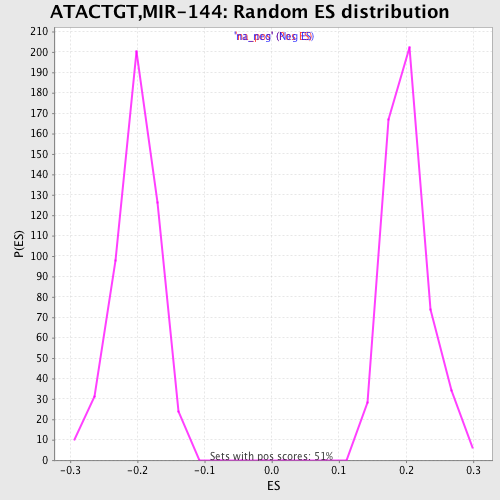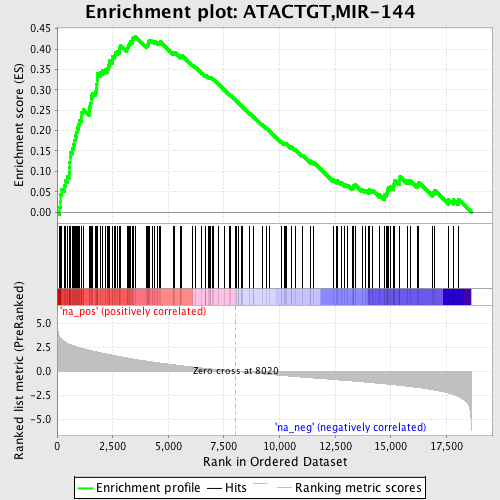
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ATACTGT,MIR-144 |
| Enrichment Score (ES) | 0.4317521 |
| Normalized Enrichment Score (NES) | 2.1510134 |
| Nominal p-value | 0.0 |
| FDR q-value | 6.8551395E-4 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | APPBP2 | 106 | 3.674 | 0.0120 | Yes | ||
| 2 | SS18 | 140 | 3.521 | 0.0273 | Yes | ||
| 3 | PPP3R1 | 143 | 3.514 | 0.0442 | Yes | ||
| 4 | HSF2 | 216 | 3.301 | 0.0563 | Yes | ||
| 5 | CLK1 | 319 | 3.127 | 0.0659 | Yes | ||
| 6 | NARG1 | 373 | 3.040 | 0.0777 | Yes | ||
| 7 | ARID2 | 468 | 2.918 | 0.0867 | Yes | ||
| 8 | GLCCI1 | 538 | 2.834 | 0.0967 | Yes | ||
| 9 | SIN3A | 566 | 2.813 | 0.1088 | Yes | ||
| 10 | ITSN2 | 574 | 2.802 | 0.1220 | Yes | ||
| 11 | MARCKS | 580 | 2.799 | 0.1353 | Yes | ||
| 12 | STAG1 | 617 | 2.762 | 0.1467 | Yes | ||
| 13 | CUGBP2 | 695 | 2.698 | 0.1556 | Yes | ||
| 14 | KPNA1 | 728 | 2.676 | 0.1668 | Yes | ||
| 15 | CDH2 | 787 | 2.624 | 0.1764 | Yes | ||
| 16 | ACSL4 | 814 | 2.604 | 0.1875 | Yes | ||
| 17 | HNRPU | 880 | 2.564 | 0.1964 | Yes | ||
| 18 | NFE2L2 | 900 | 2.550 | 0.2077 | Yes | ||
| 19 | SLC23A2 | 980 | 2.496 | 0.2155 | Yes | ||
| 20 | MTPN | 1016 | 2.477 | 0.2256 | Yes | ||
| 21 | ZFX | 1088 | 2.430 | 0.2335 | Yes | ||
| 22 | PFKFB2 | 1113 | 2.412 | 0.2439 | Yes | ||
| 23 | HAT1 | 1163 | 2.382 | 0.2528 | Yes | ||
| 24 | RBM12 | 1440 | 2.216 | 0.2486 | Yes | ||
| 25 | ARID1A | 1460 | 2.208 | 0.2582 | Yes | ||
| 26 | SON | 1486 | 2.195 | 0.2675 | Yes | ||
| 27 | UBE2D3 | 1526 | 2.167 | 0.2758 | Yes | ||
| 28 | PAFAH1B1 | 1532 | 2.165 | 0.2860 | Yes | ||
| 29 | PABPN1 | 1609 | 2.123 | 0.2922 | Yes | ||
| 30 | RNF111 | 1741 | 2.055 | 0.2951 | Yes | ||
| 31 | MAPK6 | 1757 | 2.048 | 0.3042 | Yes | ||
| 32 | CCNE2 | 1766 | 2.041 | 0.3136 | Yes | ||
| 33 | EIF4G2 | 1793 | 2.030 | 0.3220 | Yes | ||
| 34 | PDE4D | 1800 | 2.027 | 0.3315 | Yes | ||
| 35 | PKNOX1 | 1817 | 2.020 | 0.3404 | Yes | ||
| 36 | UBE2D2 | 1956 | 1.936 | 0.3423 | Yes | ||
| 37 | CCNK | 2044 | 1.893 | 0.3467 | Yes | ||
| 38 | SUCLA2 | 2155 | 1.837 | 0.3497 | Yes | ||
| 39 | ZIC4 | 2278 | 1.782 | 0.3517 | Yes | ||
| 40 | NIN | 2301 | 1.770 | 0.3591 | Yes | ||
| 41 | FNDC3A | 2352 | 1.741 | 0.3648 | Yes | ||
| 42 | PHTF2 | 2361 | 1.734 | 0.3727 | Yes | ||
| 43 | ARRDC3 | 2496 | 1.680 | 0.3736 | Yes | ||
| 44 | ATP2B2 | 2501 | 1.679 | 0.3815 | Yes | ||
| 45 | TFRC | 2578 | 1.645 | 0.3853 | Yes | ||
| 46 | GSPT1 | 2613 | 1.629 | 0.3914 | Yes | ||
| 47 | ACBD3 | 2718 | 1.579 | 0.3934 | Yes | ||
| 48 | QKI | 2799 | 1.540 | 0.3965 | Yes | ||
| 49 | ARFGEF1 | 2817 | 1.532 | 0.4030 | Yes | ||
| 50 | PUM1 | 2840 | 1.521 | 0.4092 | Yes | ||
| 51 | RHOBTB1 | 3141 | 1.396 | 0.3997 | Yes | ||
| 52 | CDH11 | 3180 | 1.378 | 0.4043 | Yes | ||
| 53 | CAPZA1 | 3201 | 1.370 | 0.4098 | Yes | ||
| 54 | PPP1R16B | 3242 | 1.351 | 0.4142 | Yes | ||
| 55 | FUSIP1 | 3288 | 1.334 | 0.4182 | Yes | ||
| 56 | GDF10 | 3369 | 1.308 | 0.4202 | Yes | ||
| 57 | ALS2 | 3387 | 1.300 | 0.4256 | Yes | ||
| 58 | NR3C1 | 3444 | 1.276 | 0.4287 | Yes | ||
| 59 | FBXO32 | 3501 | 1.255 | 0.4318 | Yes | ||
| 60 | CBX4 | 4001 | 1.065 | 0.4099 | No | ||
| 61 | PTP4A1 | 4082 | 1.036 | 0.4106 | No | ||
| 62 | PALM2 | 4099 | 1.031 | 0.4147 | No | ||
| 63 | SORCS3 | 4121 | 1.023 | 0.4185 | No | ||
| 64 | GATA3 | 4156 | 1.016 | 0.4216 | No | ||
| 65 | EIF5 | 4277 | 0.978 | 0.4198 | No | ||
| 66 | SLC7A11 | 4396 | 0.939 | 0.4180 | No | ||
| 67 | WTAP | 4510 | 0.905 | 0.4162 | No | ||
| 68 | PURA | 4614 | 0.866 | 0.4148 | No | ||
| 69 | FBXL3 | 4630 | 0.860 | 0.4182 | No | ||
| 70 | FOSB | 5227 | 0.687 | 0.3892 | No | ||
| 71 | SLC12A2 | 5236 | 0.685 | 0.3921 | No | ||
| 72 | PLAG1 | 5286 | 0.670 | 0.3927 | No | ||
| 73 | FOXO1A | 5558 | 0.592 | 0.3809 | No | ||
| 74 | CRSP2 | 5562 | 0.591 | 0.3836 | No | ||
| 75 | NGFRAP1 | 5612 | 0.576 | 0.3837 | No | ||
| 76 | CBX1 | 6075 | 0.458 | 0.3609 | No | ||
| 77 | VKORC1L1 | 6204 | 0.422 | 0.3560 | No | ||
| 78 | AHCY | 6503 | 0.349 | 0.3415 | No | ||
| 79 | FAM60A | 6655 | 0.315 | 0.3349 | No | ||
| 80 | CCT2 | 6665 | 0.312 | 0.3359 | No | ||
| 81 | BRPF1 | 6686 | 0.306 | 0.3363 | No | ||
| 82 | PTPN9 | 6822 | 0.273 | 0.3303 | No | ||
| 83 | PCDH18 | 6839 | 0.270 | 0.3308 | No | ||
| 84 | PANK1 | 6882 | 0.261 | 0.3297 | No | ||
| 85 | MYT1 | 6900 | 0.254 | 0.3301 | No | ||
| 86 | PAX3 | 6985 | 0.235 | 0.3266 | No | ||
| 87 | LIFR | 7025 | 0.226 | 0.3256 | No | ||
| 88 | CAV2 | 7254 | 0.175 | 0.3141 | No | ||
| 89 | HNRPF | 7505 | 0.115 | 0.3011 | No | ||
| 90 | FBXW11 | 7737 | 0.066 | 0.2890 | No | ||
| 91 | RGS16 | 7794 | 0.055 | 0.2862 | No | ||
| 92 | UBE4A | 8006 | 0.003 | 0.2748 | No | ||
| 93 | SP4 | 8076 | -0.012 | 0.2711 | No | ||
| 94 | ELL2 | 8163 | -0.031 | 0.2666 | No | ||
| 95 | RIN2 | 8165 | -0.031 | 0.2667 | No | ||
| 96 | SMARCA1 | 8272 | -0.053 | 0.2612 | No | ||
| 97 | PPARA | 8273 | -0.053 | 0.2615 | No | ||
| 98 | ZCCHC2 | 8334 | -0.069 | 0.2585 | No | ||
| 99 | MSX1 | 8669 | -0.138 | 0.2411 | No | ||
| 100 | ESRRG | 8806 | -0.163 | 0.2345 | No | ||
| 101 | FMN2 | 8848 | -0.174 | 0.2332 | No | ||
| 102 | WIF1 | 9209 | -0.254 | 0.2149 | No | ||
| 103 | IDH2 | 9398 | -0.294 | 0.2061 | No | ||
| 104 | SLITRK4 | 9527 | -0.325 | 0.2008 | No | ||
| 105 | LGR4 | 10085 | -0.430 | 0.1727 | No | ||
| 106 | TBX1 | 10221 | -0.458 | 0.1676 | No | ||
| 107 | MYO1E | 10274 | -0.466 | 0.1670 | No | ||
| 108 | ST6GALNAC3 | 10291 | -0.469 | 0.1684 | No | ||
| 109 | RFX4 | 10515 | -0.511 | 0.1588 | No | ||
| 110 | ST18 | 10534 | -0.515 | 0.1603 | No | ||
| 111 | SFRP1 | 10710 | -0.547 | 0.1535 | No | ||
| 112 | NRP2 | 11040 | -0.612 | 0.1386 | No | ||
| 113 | CD28 | 11391 | -0.675 | 0.1230 | No | ||
| 114 | SEMA6A | 11405 | -0.678 | 0.1255 | No | ||
| 115 | GCNT2 | 11545 | -0.702 | 0.1214 | No | ||
| 116 | BACH2 | 12401 | -0.851 | 0.0792 | No | ||
| 117 | RARB | 12546 | -0.879 | 0.0757 | No | ||
| 118 | ANK2 | 12595 | -0.888 | 0.0774 | No | ||
| 119 | ARID5B | 12768 | -0.920 | 0.0725 | No | ||
| 120 | MEIS1 | 12937 | -0.952 | 0.0680 | No | ||
| 121 | CXCL12 | 13053 | -0.972 | 0.0665 | No | ||
| 122 | STARD8 | 13275 | -1.015 | 0.0594 | No | ||
| 123 | TTN | 13315 | -1.022 | 0.0622 | No | ||
| 124 | EDAR | 13335 | -1.027 | 0.0662 | No | ||
| 125 | NEDD4 | 13400 | -1.039 | 0.0677 | No | ||
| 126 | ALDH1A3 | 13711 | -1.097 | 0.0563 | No | ||
| 127 | SNAP91 | 13871 | -1.126 | 0.0531 | No | ||
| 128 | PTGFRN | 14010 | -1.154 | 0.0512 | No | ||
| 129 | CPEB2 | 14049 | -1.163 | 0.0548 | No | ||
| 130 | HDHD2 | 14166 | -1.189 | 0.0542 | No | ||
| 131 | PNRC1 | 14494 | -1.263 | 0.0426 | No | ||
| 132 | MYST3 | 14713 | -1.304 | 0.0371 | No | ||
| 133 | DMD | 14733 | -1.309 | 0.0424 | No | ||
| 134 | ETS1 | 14799 | -1.325 | 0.0453 | No | ||
| 135 | AGRN | 14831 | -1.331 | 0.0501 | No | ||
| 136 | LHX2 | 14847 | -1.335 | 0.0557 | No | ||
| 137 | JPH1 | 14894 | -1.346 | 0.0598 | No | ||
| 138 | WDFY3 | 14976 | -1.369 | 0.0620 | No | ||
| 139 | CPEB3 | 15108 | -1.397 | 0.0617 | No | ||
| 140 | NR2F2 | 15134 | -1.401 | 0.0671 | No | ||
| 141 | PAPPA | 15172 | -1.409 | 0.0719 | No | ||
| 142 | CALCRL | 15180 | -1.410 | 0.0783 | No | ||
| 143 | PIM1 | 15392 | -1.461 | 0.0740 | No | ||
| 144 | HOXA10 | 15394 | -1.462 | 0.0810 | No | ||
| 145 | SMPD3 | 15411 | -1.467 | 0.0872 | No | ||
| 146 | ATXN1 | 15736 | -1.552 | 0.0772 | No | ||
| 147 | TRIB1 | 15881 | -1.594 | 0.0771 | No | ||
| 148 | PDE7B | 16197 | -1.681 | 0.0682 | No | ||
| 149 | ATP5G2 | 16259 | -1.698 | 0.0731 | No | ||
| 150 | FMR1 | 16887 | -1.931 | 0.0485 | No | ||
| 151 | SEMA6D | 16975 | -1.966 | 0.0533 | No | ||
| 152 | PTHLH | 17605 | -2.275 | 0.0302 | No | ||
| 153 | DGCR2 | 17798 | -2.423 | 0.0315 | No | ||
| 154 | DLG5 | 18030 | -2.628 | 0.0317 | No |