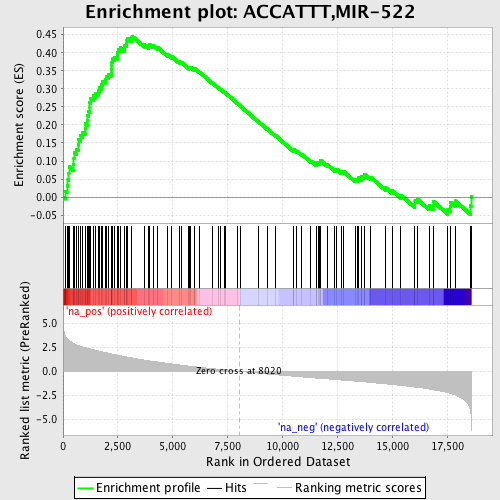
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ACCATTT,MIR-522 |
| Enrichment Score (ES) | 0.44603986 |
| Normalized Enrichment Score (NES) | 2.0973935 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.125228E-4 |
| FWER p-Value | 0.0080 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EHMT1 | 98 | 3.704 | 0.0165 | Yes | ||
| 2 | AKAP9 | 178 | 3.420 | 0.0323 | Yes | ||
| 3 | MECP2 | 220 | 3.287 | 0.0494 | Yes | ||
| 4 | GOLGA1 | 265 | 3.212 | 0.0659 | Yes | ||
| 5 | PRPF39 | 269 | 3.203 | 0.0845 | Yes | ||
| 6 | ARID2 | 468 | 2.918 | 0.0910 | Yes | ||
| 7 | KPNB1 | 493 | 2.884 | 0.1066 | Yes | ||
| 8 | TOP1 | 502 | 2.872 | 0.1230 | Yes | ||
| 9 | STAG1 | 617 | 2.762 | 0.1331 | Yes | ||
| 10 | CUGBP2 | 695 | 2.698 | 0.1448 | Yes | ||
| 11 | VAPA | 706 | 2.692 | 0.1601 | Yes | ||
| 12 | MBNL1 | 798 | 2.617 | 0.1705 | Yes | ||
| 13 | NFE2L2 | 900 | 2.550 | 0.1800 | Yes | ||
| 14 | MYEF2 | 1005 | 2.483 | 0.1890 | Yes | ||
| 15 | HNRPM | 1012 | 2.479 | 0.2032 | Yes | ||
| 16 | KIF23 | 1105 | 2.417 | 0.2125 | Yes | ||
| 17 | TOPORS | 1122 | 2.407 | 0.2257 | Yes | ||
| 18 | SYNCRIP | 1162 | 2.383 | 0.2376 | Yes | ||
| 19 | TMEM2 | 1209 | 2.352 | 0.2490 | Yes | ||
| 20 | KRAS | 1213 | 2.349 | 0.2626 | Yes | ||
| 21 | ELAVL1 | 1266 | 2.321 | 0.2734 | Yes | ||
| 22 | GPIAP1 | 1373 | 2.255 | 0.2809 | Yes | ||
| 23 | UBE2N | 1493 | 2.188 | 0.2874 | Yes | ||
| 24 | RBMX | 1593 | 2.135 | 0.2946 | Yes | ||
| 25 | FEM1C | 1661 | 2.094 | 0.3032 | Yes | ||
| 26 | LRCH2 | 1740 | 2.055 | 0.3111 | Yes | ||
| 27 | CUL1 | 1780 | 2.035 | 0.3209 | Yes | ||
| 28 | GAB2 | 1917 | 1.959 | 0.3251 | Yes | ||
| 29 | HNRPK | 1954 | 1.937 | 0.3345 | Yes | ||
| 30 | ING3 | 2068 | 1.878 | 0.3395 | Yes | ||
| 31 | YWHAZ | 2185 | 1.824 | 0.3439 | Yes | ||
| 32 | MAN1A2 | 2208 | 1.816 | 0.3534 | Yes | ||
| 33 | ATP1B1 | 2217 | 1.812 | 0.3636 | Yes | ||
| 34 | UBE2L3 | 2222 | 1.809 | 0.3740 | Yes | ||
| 35 | RSBN1 | 2256 | 1.791 | 0.3827 | Yes | ||
| 36 | UBE3A | 2353 | 1.740 | 0.3878 | Yes | ||
| 37 | CDV3 | 2464 | 1.693 | 0.3918 | Yes | ||
| 38 | IVNS1ABP | 2484 | 1.685 | 0.4007 | Yes | ||
| 39 | SLC7A6 | 2505 | 1.678 | 0.4094 | Yes | ||
| 40 | ZC3H7A | 2607 | 1.631 | 0.4136 | Yes | ||
| 41 | CPD | 2801 | 1.539 | 0.4122 | Yes | ||
| 42 | GNAO1 | 2811 | 1.535 | 0.4207 | Yes | ||
| 43 | RTF1 | 2891 | 1.499 | 0.4252 | Yes | ||
| 44 | TBC1D8 | 2908 | 1.493 | 0.4331 | Yes | ||
| 45 | MYH10 | 2933 | 1.483 | 0.4406 | Yes | ||
| 46 | CASKIN1 | 3120 | 1.404 | 0.4388 | Yes | ||
| 47 | CDS2 | 3138 | 1.396 | 0.4460 | Yes | ||
| 48 | PTPN11 | 3701 | 1.174 | 0.4226 | No | ||
| 49 | NPAT | 3891 | 1.106 | 0.4189 | No | ||
| 50 | DAG1 | 3935 | 1.089 | 0.4229 | No | ||
| 51 | PHYHIPL | 4102 | 1.030 | 0.4200 | No | ||
| 52 | LRP1B | 4293 | 0.970 | 0.4154 | No | ||
| 53 | NR4A3 | 4766 | 0.823 | 0.3948 | No | ||
| 54 | AKAP1 | 4951 | 0.770 | 0.3893 | No | ||
| 55 | PLAG1 | 5286 | 0.670 | 0.3752 | No | ||
| 56 | DYRK1A | 5380 | 0.637 | 0.3740 | No | ||
| 57 | SLC39A9 | 5736 | 0.541 | 0.3579 | No | ||
| 58 | YWHAQ | 5777 | 0.530 | 0.3589 | No | ||
| 59 | PRKG1 | 5811 | 0.522 | 0.3602 | No | ||
| 60 | FOXP1 | 5973 | 0.483 | 0.3543 | No | ||
| 61 | RBM22 | 5997 | 0.477 | 0.3559 | No | ||
| 62 | TRPM7 | 6215 | 0.419 | 0.3466 | No | ||
| 63 | NFKBIZ | 6807 | 0.277 | 0.3163 | No | ||
| 64 | BHLHB5 | 7063 | 0.218 | 0.3038 | No | ||
| 65 | ENC1 | 7182 | 0.195 | 0.2986 | No | ||
| 66 | CBLL1 | 7365 | 0.149 | 0.2896 | No | ||
| 67 | MAFB | 7391 | 0.145 | 0.2891 | No | ||
| 68 | CCND2 | 7931 | 0.024 | 0.2601 | No | ||
| 69 | SP4 | 8076 | -0.012 | 0.2524 | No | ||
| 70 | PANK2 | 8892 | -0.184 | 0.2095 | No | ||
| 71 | TLX1 | 9293 | -0.276 | 0.1895 | No | ||
| 72 | ARHGAP5 | 9685 | -0.356 | 0.1704 | No | ||
| 73 | UBE2J1 | 10483 | -0.505 | 0.1303 | No | ||
| 74 | WNK3 | 10502 | -0.509 | 0.1324 | No | ||
| 75 | CEBPA | 10653 | -0.537 | 0.1274 | No | ||
| 76 | RNF2 | 10865 | -0.575 | 0.1194 | No | ||
| 77 | JUN | 11297 | -0.660 | 0.1000 | No | ||
| 78 | AQP9 | 11529 | -0.700 | 0.0916 | No | ||
| 79 | GPM6B | 11543 | -0.702 | 0.0950 | No | ||
| 80 | NAB1 | 11621 | -0.715 | 0.0951 | No | ||
| 81 | HBP1 | 11689 | -0.724 | 0.0957 | No | ||
| 82 | MEF2D | 11709 | -0.727 | 0.0989 | No | ||
| 83 | TNPO2 | 11727 | -0.729 | 0.1023 | No | ||
| 84 | CSNK1A1 | 12034 | -0.787 | 0.0904 | No | ||
| 85 | WDR26 | 12386 | -0.849 | 0.0764 | No | ||
| 86 | ARMCX2 | 12471 | -0.865 | 0.0770 | No | ||
| 87 | PERP | 12704 | -0.907 | 0.0698 | No | ||
| 88 | ARID5B | 12768 | -0.920 | 0.0717 | No | ||
| 89 | TTN | 13315 | -1.022 | 0.0482 | No | ||
| 90 | PLK2 | 13423 | -1.043 | 0.0486 | No | ||
| 91 | ETV1 | 13444 | -1.046 | 0.0537 | No | ||
| 92 | DOCK7 | 13587 | -1.076 | 0.0523 | No | ||
| 93 | CNTFR | 13605 | -1.080 | 0.0577 | No | ||
| 94 | HOXB5 | 13718 | -1.098 | 0.0581 | No | ||
| 95 | SALL1 | 13747 | -1.103 | 0.0631 | No | ||
| 96 | PTGFRN | 14010 | -1.154 | 0.0557 | No | ||
| 97 | SOX9 | 14712 | -1.304 | 0.0255 | No | ||
| 98 | ZFP36L2 | 15010 | -1.375 | 0.0175 | No | ||
| 99 | CTBP1 | 15395 | -1.462 | 0.0054 | No | ||
| 100 | TCFL5 | 16031 | -1.641 | -0.0193 | No | ||
| 101 | FLRT3 | 16037 | -1.642 | -0.0099 | No | ||
| 102 | CAPN3 | 16137 | -1.666 | -0.0055 | No | ||
| 103 | CSPG5 | 16681 | -1.848 | -0.0240 | No | ||
| 104 | ATRX | 16863 | -1.921 | -0.0225 | No | ||
| 105 | FMR1 | 16887 | -1.931 | -0.0124 | No | ||
| 106 | APH1A | 17539 | -2.238 | -0.0344 | No | ||
| 107 | PTPN1 | 17641 | -2.295 | -0.0264 | No | ||
| 108 | FZD2 | 17659 | -2.314 | -0.0137 | No | ||
| 109 | WDTC1 | 17870 | -2.471 | -0.0105 | No | ||
| 110 | SH3BP5 | 18566 | -4.110 | -0.0240 | No | ||
| 111 | CRIM1 | 18594 | -4.529 | 0.0012 | No |
