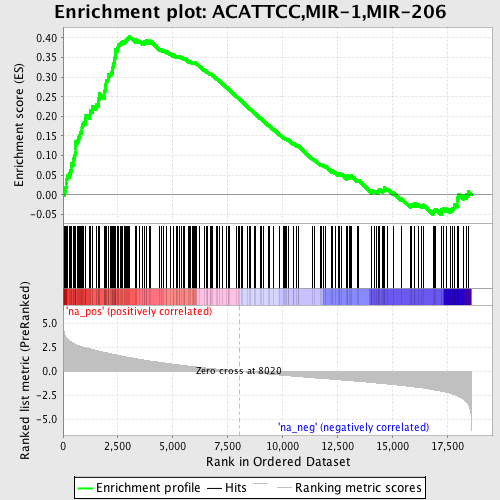
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ACATTCC,MIR-1,MIR-206 |
| Enrichment Score (ES) | 0.40479922 |
| Normalized Enrichment Score (NES) | 2.101072 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.075258E-4 |
| FWER p-Value | 0.0080 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BAG4 | 59 | 3.972 | 0.0098 | Yes | ||
| 2 | AP1G1 | 114 | 3.623 | 0.0188 | Yes | ||
| 3 | SS18 | 140 | 3.521 | 0.0290 | Yes | ||
| 4 | RNF138 | 152 | 3.488 | 0.0399 | Yes | ||
| 5 | HOXB4 | 180 | 3.412 | 0.0496 | Yes | ||
| 6 | SFRS1 | 283 | 3.183 | 0.0545 | Yes | ||
| 7 | TPM3 | 329 | 3.115 | 0.0623 | Yes | ||
| 8 | PDCD10 | 377 | 3.036 | 0.0697 | Yes | ||
| 9 | UBQLN1 | 384 | 3.025 | 0.0793 | Yes | ||
| 10 | ARID2 | 468 | 2.918 | 0.0844 | Yes | ||
| 11 | RASA1 | 476 | 2.908 | 0.0936 | Yes | ||
| 12 | EIF4E | 531 | 2.842 | 0.1000 | Yes | ||
| 13 | FRAS1 | 544 | 2.831 | 0.1086 | Yes | ||
| 14 | DHX15 | 558 | 2.822 | 0.1172 | Yes | ||
| 15 | PPARBP | 563 | 2.818 | 0.1262 | Yes | ||
| 16 | C1GALT1 | 572 | 2.805 | 0.1350 | Yes | ||
| 17 | OGT | 665 | 2.718 | 0.1389 | Yes | ||
| 18 | SNX2 | 708 | 2.691 | 0.1455 | Yes | ||
| 19 | SFRS3 | 745 | 2.665 | 0.1523 | Yes | ||
| 20 | DNAJC5 | 769 | 2.642 | 0.1597 | Yes | ||
| 21 | SMYD4 | 856 | 2.576 | 0.1635 | Yes | ||
| 22 | RNGTT | 859 | 2.575 | 0.1718 | Yes | ||
| 23 | HNRPU | 880 | 2.564 | 0.1792 | Yes | ||
| 24 | SLC39A10 | 949 | 2.516 | 0.1837 | Yes | ||
| 25 | EAF1 | 1017 | 2.476 | 0.1882 | Yes | ||
| 26 | NCL | 1029 | 2.462 | 0.1957 | Yes | ||
| 27 | KLF13 | 1040 | 2.454 | 0.2032 | Yes | ||
| 28 | ABCB7 | 1186 | 2.367 | 0.2031 | Yes | ||
| 29 | CNN3 | 1225 | 2.343 | 0.2088 | Yes | ||
| 30 | HMGCR | 1245 | 2.332 | 0.2154 | Yes | ||
| 31 | RNUXA | 1333 | 2.278 | 0.2182 | Yes | ||
| 32 | ANP32B | 1341 | 2.275 | 0.2252 | Yes | ||
| 33 | PUM2 | 1500 | 2.184 | 0.2238 | Yes | ||
| 34 | PAFAH1B1 | 1532 | 2.165 | 0.2293 | Yes | ||
| 35 | VAMP4 | 1601 | 2.129 | 0.2325 | Yes | ||
| 36 | HSPD1 | 1622 | 2.116 | 0.2384 | Yes | ||
| 37 | CLCN3 | 1631 | 2.113 | 0.2449 | Yes | ||
| 38 | PHF6 | 1646 | 2.099 | 0.2511 | Yes | ||
| 39 | STARD7 | 1650 | 2.098 | 0.2578 | Yes | ||
| 40 | ATP6V1A | 1880 | 1.981 | 0.2518 | Yes | ||
| 41 | NUP50 | 1881 | 1.980 | 0.2583 | Yes | ||
| 42 | TTC7B | 1883 | 1.980 | 0.2648 | Yes | ||
| 43 | SFRS10 | 1915 | 1.959 | 0.2695 | Yes | ||
| 44 | DNAJC13 | 1922 | 1.955 | 0.2756 | Yes | ||
| 45 | MATR3 | 1929 | 1.952 | 0.2817 | Yes | ||
| 46 | HNRPK | 1954 | 1.937 | 0.2868 | Yes | ||
| 47 | SMARCA4 | 1963 | 1.934 | 0.2927 | Yes | ||
| 48 | AKAP12 | 2045 | 1.893 | 0.2945 | Yes | ||
| 49 | CDC42 | 2048 | 1.892 | 0.3006 | Yes | ||
| 50 | TBP | 2064 | 1.880 | 0.3060 | Yes | ||
| 51 | OTX2 | 2137 | 1.843 | 0.3081 | Yes | ||
| 52 | YWHAZ | 2185 | 1.824 | 0.3116 | Yes | ||
| 53 | GNPDA2 | 2236 | 1.801 | 0.3148 | Yes | ||
| 54 | RSBN1 | 2256 | 1.791 | 0.3196 | Yes | ||
| 55 | USP33 | 2265 | 1.789 | 0.3250 | Yes | ||
| 56 | ZIC4 | 2278 | 1.782 | 0.3302 | Yes | ||
| 57 | AZIN1 | 2305 | 1.768 | 0.3346 | Yes | ||
| 58 | POLDIP3 | 2321 | 1.758 | 0.3396 | Yes | ||
| 59 | FNDC3A | 2352 | 1.741 | 0.3437 | Yes | ||
| 60 | SEC63 | 2360 | 1.735 | 0.3490 | Yes | ||
| 61 | FBXW7 | 2368 | 1.732 | 0.3543 | Yes | ||
| 62 | ARF3 | 2382 | 1.728 | 0.3593 | Yes | ||
| 63 | SMAP1 | 2385 | 1.727 | 0.3649 | Yes | ||
| 64 | ARCN1 | 2386 | 1.727 | 0.3705 | Yes | ||
| 65 | MMD | 2465 | 1.693 | 0.3718 | Yes | ||
| 66 | ARRDC3 | 2496 | 1.680 | 0.3757 | Yes | ||
| 67 | SLC7A6 | 2505 | 1.678 | 0.3808 | Yes | ||
| 68 | BCL7A | 2544 | 1.659 | 0.3842 | Yes | ||
| 69 | DDX5 | 2595 | 1.638 | 0.3869 | Yes | ||
| 70 | EIF2C1 | 2674 | 1.597 | 0.3879 | Yes | ||
| 71 | AP3D1 | 2724 | 1.576 | 0.3904 | Yes | ||
| 72 | QKI | 2799 | 1.540 | 0.3914 | Yes | ||
| 73 | PICALM | 2847 | 1.519 | 0.3938 | Yes | ||
| 74 | RRBP1 | 2909 | 1.492 | 0.3954 | Yes | ||
| 75 | UST | 2919 | 1.489 | 0.3998 | Yes | ||
| 76 | SULF1 | 3000 | 1.452 | 0.4003 | Yes | ||
| 77 | BPNT1 | 3005 | 1.449 | 0.4048 | Yes | ||
| 78 | UBE2H | 3301 | 1.330 | 0.3931 | No | ||
| 79 | GPD2 | 3328 | 1.322 | 0.3961 | No | ||
| 80 | LIN7C | 3481 | 1.262 | 0.3919 | No | ||
| 81 | HIAT1 | 3629 | 1.203 | 0.3879 | No | ||
| 82 | PFTK1 | 3719 | 1.166 | 0.3869 | No | ||
| 83 | HMGN1 | 3721 | 1.165 | 0.3907 | No | ||
| 84 | TIMP3 | 3801 | 1.139 | 0.3901 | No | ||
| 85 | NCK2 | 3813 | 1.136 | 0.3933 | No | ||
| 86 | BLCAP | 3945 | 1.086 | 0.3897 | No | ||
| 87 | MEOX2 | 3961 | 1.079 | 0.3924 | No | ||
| 88 | E2F5 | 4408 | 0.936 | 0.3713 | No | ||
| 89 | DGKG | 4492 | 0.910 | 0.3697 | No | ||
| 90 | BDNF | 4590 | 0.876 | 0.3674 | No | ||
| 91 | IGF1 | 4695 | 0.840 | 0.3645 | No | ||
| 92 | BSN | 4717 | 0.834 | 0.3661 | No | ||
| 93 | ZFP36L1 | 4874 | 0.797 | 0.3602 | No | ||
| 94 | AKAP11 | 5015 | 0.752 | 0.3551 | No | ||
| 95 | FGF14 | 5028 | 0.748 | 0.3569 | No | ||
| 96 | ANXA2 | 5154 | 0.707 | 0.3524 | No | ||
| 97 | ARHGEF18 | 5183 | 0.699 | 0.3532 | No | ||
| 98 | FOSB | 5227 | 0.687 | 0.3531 | No | ||
| 99 | FBXO33 | 5296 | 0.667 | 0.3516 | No | ||
| 100 | CPEB1 | 5309 | 0.664 | 0.3531 | No | ||
| 101 | SLC35F1 | 5415 | 0.628 | 0.3495 | No | ||
| 102 | MAPKBP1 | 5474 | 0.615 | 0.3483 | No | ||
| 103 | FN1 | 5535 | 0.597 | 0.3470 | No | ||
| 104 | TGFBR3 | 5551 | 0.593 | 0.3482 | No | ||
| 105 | SETBP1 | 5734 | 0.541 | 0.3401 | No | ||
| 106 | YWHAQ | 5777 | 0.530 | 0.3395 | No | ||
| 107 | ADPGK | 5793 | 0.527 | 0.3404 | No | ||
| 108 | EFNB2 | 5884 | 0.503 | 0.3372 | No | ||
| 109 | MKL1 | 5943 | 0.490 | 0.3356 | No | ||
| 110 | SLC8A2 | 5958 | 0.486 | 0.3365 | No | ||
| 111 | FOXP1 | 5973 | 0.483 | 0.3373 | No | ||
| 112 | EDN1 | 6015 | 0.473 | 0.3366 | No | ||
| 113 | NCOA3 | 6101 | 0.452 | 0.3335 | No | ||
| 114 | RSBN1L | 6227 | 0.415 | 0.3281 | No | ||
| 115 | BSCL2 | 6434 | 0.367 | 0.3181 | No | ||
| 116 | SLC25A22 | 6519 | 0.346 | 0.3147 | No | ||
| 117 | SP2 | 6582 | 0.332 | 0.3124 | No | ||
| 118 | CBL | 6592 | 0.331 | 0.3130 | No | ||
| 119 | MAL2 | 6699 | 0.303 | 0.3082 | No | ||
| 120 | ATP7A | 6718 | 0.299 | 0.3082 | No | ||
| 121 | COL4A3 | 6766 | 0.288 | 0.3066 | No | ||
| 122 | CORO1C | 6828 | 0.273 | 0.3042 | No | ||
| 123 | PAX3 | 6985 | 0.235 | 0.2965 | No | ||
| 124 | OLFML2A | 7035 | 0.224 | 0.2945 | No | ||
| 125 | DSCR1L1 | 7127 | 0.207 | 0.2903 | No | ||
| 126 | SFRS9 | 7264 | 0.172 | 0.2835 | No | ||
| 127 | CLTC | 7450 | 0.132 | 0.2738 | No | ||
| 128 | PPIB | 7521 | 0.113 | 0.2704 | No | ||
| 129 | TLOC1 | 7549 | 0.109 | 0.2693 | No | ||
| 130 | GDAP1L1 | 7577 | 0.102 | 0.2682 | No | ||
| 131 | SPRED1 | 7924 | 0.025 | 0.2494 | No | ||
| 132 | UBE4A | 8006 | 0.003 | 0.2450 | No | ||
| 133 | MAP4K3 | 8043 | -0.003 | 0.2431 | No | ||
| 134 | SLC25A25 | 8123 | -0.022 | 0.2389 | No | ||
| 135 | CDK9 | 8160 | -0.030 | 0.2370 | No | ||
| 136 | GJA1 | 8400 | -0.082 | 0.2243 | No | ||
| 137 | KCTD13 | 8493 | -0.104 | 0.2196 | No | ||
| 138 | TFEC | 8535 | -0.113 | 0.2178 | No | ||
| 139 | MIPOL1 | 8721 | -0.148 | 0.2082 | No | ||
| 140 | NGFR | 8769 | -0.158 | 0.2062 | No | ||
| 141 | POGK | 8979 | -0.202 | 0.1955 | No | ||
| 142 | EYA4 | 9021 | -0.213 | 0.1939 | No | ||
| 143 | UBN1 | 9122 | -0.234 | 0.1893 | No | ||
| 144 | ANKRD29 | 9373 | -0.290 | 0.1766 | No | ||
| 145 | WDR6 | 9412 | -0.296 | 0.1756 | No | ||
| 146 | TGIF2 | 9608 | -0.342 | 0.1661 | No | ||
| 147 | VAMP2 | 9881 | -0.393 | 0.1526 | No | ||
| 148 | GAS2L1 | 10051 | -0.423 | 0.1448 | No | ||
| 149 | CEBPZ | 10086 | -0.430 | 0.1444 | No | ||
| 150 | TKT | 10145 | -0.441 | 0.1426 | No | ||
| 151 | MET | 10182 | -0.448 | 0.1422 | No | ||
| 152 | WDR40B | 10264 | -0.464 | 0.1393 | No | ||
| 153 | MYO1E | 10274 | -0.466 | 0.1403 | No | ||
| 154 | NAP1L5 | 10499 | -0.508 | 0.1298 | No | ||
| 155 | RIMS4 | 10500 | -0.508 | 0.1315 | No | ||
| 156 | MAB21L1 | 10649 | -0.537 | 0.1252 | No | ||
| 157 | SFRP1 | 10710 | -0.547 | 0.1237 | No | ||
| 158 | STC2 | 10724 | -0.549 | 0.1248 | No | ||
| 159 | LRRC8A | 11356 | -0.670 | 0.0928 | No | ||
| 160 | CREM | 11466 | -0.689 | 0.0891 | No | ||
| 161 | TNPO2 | 11727 | -0.729 | 0.0774 | No | ||
| 162 | VGLL4 | 11792 | -0.738 | 0.0763 | No | ||
| 163 | SLC7A2 | 11855 | -0.753 | 0.0754 | No | ||
| 164 | HIC2 | 11960 | -0.773 | 0.0723 | No | ||
| 165 | SUHW4 | 12234 | -0.823 | 0.0602 | No | ||
| 166 | CITED2 | 12257 | -0.828 | 0.0617 | No | ||
| 167 | BACH2 | 12401 | -0.851 | 0.0567 | No | ||
| 168 | RARB | 12546 | -0.879 | 0.0518 | No | ||
| 169 | CDK6 | 12593 | -0.888 | 0.0522 | No | ||
| 170 | SLC38A3 | 12615 | -0.893 | 0.0540 | No | ||
| 171 | TMOD2 | 12699 | -0.905 | 0.0524 | No | ||
| 172 | MEIS1 | 12937 | -0.952 | 0.0427 | No | ||
| 173 | BET1 | 12945 | -0.953 | 0.0454 | No | ||
| 174 | ZBTB4 | 12952 | -0.955 | 0.0482 | No | ||
| 175 | GDF6 | 13058 | -0.973 | 0.0457 | No | ||
| 176 | CPLX2 | 13097 | -0.980 | 0.0469 | No | ||
| 177 | SNAP25 | 13123 | -0.983 | 0.0487 | No | ||
| 178 | RNF38 | 13398 | -1.039 | 0.0373 | No | ||
| 179 | GPR125 | 13482 | -1.054 | 0.0362 | No | ||
| 180 | HS3ST3B1 | 14072 | -1.166 | 0.0080 | No | ||
| 181 | MYLK | 14075 | -1.166 | 0.0118 | No | ||
| 182 | PTPRG | 14208 | -1.198 | 0.0085 | No | ||
| 183 | NR4A2 | 14288 | -1.217 | 0.0082 | No | ||
| 184 | DAAM1 | 14364 | -1.235 | 0.0082 | No | ||
| 185 | TBC1D15 | 14388 | -1.240 | 0.0110 | No | ||
| 186 | FZD7 | 14426 | -1.249 | 0.0131 | No | ||
| 187 | SYNJ2 | 14548 | -1.275 | 0.0107 | No | ||
| 188 | HIVEP2 | 14613 | -1.287 | 0.0115 | No | ||
| 189 | YPEL2 | 14642 | -1.293 | 0.0142 | No | ||
| 190 | SLC37A3 | 14655 | -1.295 | 0.0178 | No | ||
| 191 | ETS1 | 14799 | -1.325 | 0.0144 | No | ||
| 192 | MAP1A | 15060 | -1.386 | 0.0048 | No | ||
| 193 | SRGAP2 | 15403 | -1.466 | -0.0090 | No | ||
| 194 | ADAR | 15835 | -1.582 | -0.0272 | No | ||
| 195 | RIT2 | 15891 | -1.597 | -0.0249 | No | ||
| 196 | ANXA4 | 16007 | -1.633 | -0.0258 | No | ||
| 197 | RSPO3 | 16027 | -1.639 | -0.0215 | No | ||
| 198 | TRAPPC3 | 16200 | -1.683 | -0.0253 | No | ||
| 199 | CREB5 | 16342 | -1.724 | -0.0273 | No | ||
| 200 | BICD1 | 16428 | -1.754 | -0.0262 | No | ||
| 201 | RNF13 | 16859 | -1.920 | -0.0432 | No | ||
| 202 | SH3BGRL3 | 16907 | -1.940 | -0.0394 | No | ||
| 203 | SEMA6D | 16975 | -1.966 | -0.0366 | No | ||
| 204 | KTN1 | 17230 | -2.074 | -0.0436 | No | ||
| 205 | TSPAN4 | 17247 | -2.082 | -0.0376 | No | ||
| 206 | GCH1 | 17322 | -2.113 | -0.0347 | No | ||
| 207 | DGKZ | 17485 | -2.208 | -0.0362 | No | ||
| 208 | TRIM2 | 17673 | -2.323 | -0.0388 | No | ||
| 209 | ACACA | 17730 | -2.366 | -0.0340 | No | ||
| 210 | NR1H3 | 17815 | -2.434 | -0.0306 | No | ||
| 211 | PIAS3 | 17855 | -2.463 | -0.0246 | No | ||
| 212 | TAGLN2 | 17980 | -2.579 | -0.0229 | No | ||
| 213 | PHC2 | 17981 | -2.579 | -0.0144 | No | ||
| 214 | MNT | 17992 | -2.589 | -0.0065 | No | ||
| 215 | GRK6 | 18038 | -2.636 | -0.0003 | No | ||
| 216 | CELSR3 | 18266 | -2.945 | -0.0029 | No | ||
| 217 | RKHD2 | 18400 | -3.218 | 0.0004 | No | ||
| 218 | SMARCB1 | 18477 | -3.434 | 0.0076 | No |
