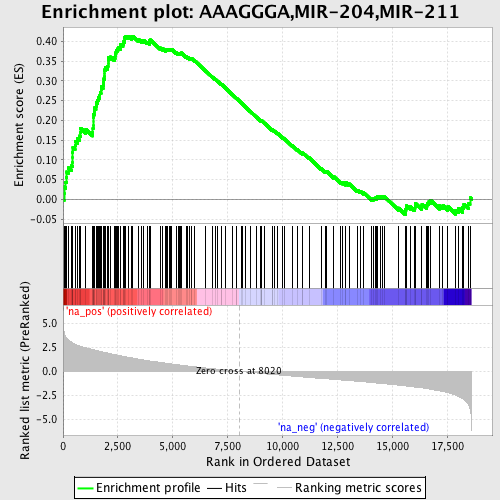
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | AAAGGGA,MIR-204,MIR-211 |
| Enrichment Score (ES) | 0.41331422 |
| Normalized Enrichment Score (NES) | 2.080936 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.9119805E-4 |
| FWER p-Value | 0.01 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | METAP1 | 43 | 4.113 | 0.0149 | Yes | ||
| 2 | TAF5 | 50 | 4.062 | 0.0315 | Yes | ||
| 3 | DNM2 | 117 | 3.612 | 0.0431 | Yes | ||
| 4 | PPP3R1 | 143 | 3.514 | 0.0564 | Yes | ||
| 5 | ELOVL6 | 171 | 3.425 | 0.0693 | Yes | ||
| 6 | BRD4 | 231 | 3.270 | 0.0797 | Yes | ||
| 7 | NARG1 | 373 | 3.040 | 0.0848 | Yes | ||
| 8 | RHOT1 | 428 | 2.961 | 0.0943 | Yes | ||
| 9 | WSB1 | 435 | 2.947 | 0.1063 | Yes | ||
| 10 | RAB1A | 439 | 2.944 | 0.1184 | Yes | ||
| 11 | DMTF1 | 447 | 2.940 | 0.1303 | Yes | ||
| 12 | FRAS1 | 544 | 2.831 | 0.1369 | Yes | ||
| 13 | SIN3A | 566 | 2.813 | 0.1476 | Yes | ||
| 14 | CRKL | 669 | 2.716 | 0.1534 | Yes | ||
| 15 | RAB14 | 746 | 2.665 | 0.1604 | Yes | ||
| 16 | CDH2 | 787 | 2.624 | 0.1692 | Yes | ||
| 17 | MBNL1 | 798 | 2.617 | 0.1796 | Yes | ||
| 18 | HS2ST1 | 1036 | 2.459 | 0.1771 | Yes | ||
| 19 | ITPR1 | 1346 | 2.271 | 0.1698 | Yes | ||
| 20 | KITLG | 1360 | 2.263 | 0.1786 | Yes | ||
| 21 | TMEM32 | 1368 | 2.259 | 0.1876 | Yes | ||
| 22 | AP3M1 | 1372 | 2.256 | 0.1969 | Yes | ||
| 23 | GPIAP1 | 1373 | 2.255 | 0.2063 | Yes | ||
| 24 | UBE2R2 | 1381 | 2.250 | 0.2154 | Yes | ||
| 25 | RAP2C | 1416 | 2.232 | 0.2229 | Yes | ||
| 26 | XRN1 | 1429 | 2.223 | 0.2315 | Yes | ||
| 27 | YTHDF3 | 1499 | 2.184 | 0.2369 | Yes | ||
| 28 | ZFYVE20 | 1517 | 2.170 | 0.2450 | Yes | ||
| 29 | ARL8B | 1576 | 2.142 | 0.2509 | Yes | ||
| 30 | SPOP | 1608 | 2.123 | 0.2581 | Yes | ||
| 31 | RPS6KA5 | 1673 | 2.089 | 0.2633 | Yes | ||
| 32 | VIL2 | 1681 | 2.085 | 0.2717 | Yes | ||
| 33 | TMOD3 | 1729 | 2.060 | 0.2777 | Yes | ||
| 34 | SIRT1 | 1739 | 2.056 | 0.2858 | Yes | ||
| 35 | KHDRBS1 | 1822 | 2.018 | 0.2898 | Yes | ||
| 36 | UHRF2 | 1844 | 2.003 | 0.2971 | Yes | ||
| 37 | BCL11B | 1857 | 1.998 | 0.3048 | Yes | ||
| 38 | YWHAG | 1884 | 1.980 | 0.3116 | Yes | ||
| 39 | MYO10 | 1891 | 1.977 | 0.3196 | Yes | ||
| 40 | TCF12 | 1894 | 1.970 | 0.3277 | Yes | ||
| 41 | DNAJC13 | 1922 | 1.955 | 0.3344 | Yes | ||
| 42 | TRIP12 | 2021 | 1.907 | 0.3371 | Yes | ||
| 43 | CDC42 | 2048 | 1.892 | 0.3436 | Yes | ||
| 44 | SEC61A2 | 2052 | 1.889 | 0.3513 | Yes | ||
| 45 | IGF2R | 2059 | 1.882 | 0.3589 | Yes | ||
| 46 | COX5A | 2169 | 1.830 | 0.3606 | Yes | ||
| 47 | ATF2 | 2330 | 1.753 | 0.3593 | Yes | ||
| 48 | FBXW7 | 2368 | 1.732 | 0.3645 | Yes | ||
| 49 | ARCN1 | 2386 | 1.727 | 0.3708 | Yes | ||
| 50 | HOOK3 | 2412 | 1.717 | 0.3766 | Yes | ||
| 51 | CCNT2 | 2466 | 1.693 | 0.3808 | Yes | ||
| 52 | RERE | 2517 | 1.672 | 0.3851 | Yes | ||
| 53 | NTRK2 | 2621 | 1.621 | 0.3863 | Yes | ||
| 54 | SOX4 | 2626 | 1.618 | 0.3929 | Yes | ||
| 55 | WDR22 | 2730 | 1.572 | 0.3939 | Yes | ||
| 56 | RPS6KA3 | 2760 | 1.562 | 0.3988 | Yes | ||
| 57 | SOCS6 | 2794 | 1.541 | 0.4035 | Yes | ||
| 58 | CPD | 2801 | 1.539 | 0.4096 | Yes | ||
| 59 | SUMO2 | 2858 | 1.516 | 0.4129 | Yes | ||
| 60 | PRRX1 | 2967 | 1.465 | 0.4132 | Yes | ||
| 61 | DHH | 3111 | 1.407 | 0.4113 | Yes | ||
| 62 | KLHL13 | 3181 | 1.378 | 0.4133 | Yes | ||
| 63 | NR3C1 | 3444 | 1.276 | 0.4045 | No | ||
| 64 | HMGA2 | 3580 | 1.219 | 0.4022 | No | ||
| 65 | RAB10 | 3674 | 1.183 | 0.4021 | No | ||
| 66 | WEE1 | 3832 | 1.131 | 0.3984 | No | ||
| 67 | DAG1 | 3935 | 1.089 | 0.3974 | No | ||
| 68 | EFNB3 | 3944 | 1.086 | 0.4015 | No | ||
| 69 | RUNX2 | 3986 | 1.071 | 0.4038 | No | ||
| 70 | NRBF2 | 4435 | 0.927 | 0.3834 | No | ||
| 71 | PPARGC1A | 4538 | 0.893 | 0.3816 | No | ||
| 72 | LRRC8D | 4664 | 0.849 | 0.3783 | No | ||
| 73 | SATB2 | 4690 | 0.842 | 0.3805 | No | ||
| 74 | MLLT3 | 4763 | 0.824 | 0.3800 | No | ||
| 75 | EPHA7 | 4832 | 0.807 | 0.3797 | No | ||
| 76 | EIF2C4 | 4897 | 0.789 | 0.3796 | No | ||
| 77 | AKAP1 | 4951 | 0.770 | 0.3799 | No | ||
| 78 | SERINC3 | 5168 | 0.704 | 0.3711 | No | ||
| 79 | CENTD1 | 5239 | 0.685 | 0.3702 | No | ||
| 80 | PLAG1 | 5286 | 0.670 | 0.3705 | No | ||
| 81 | BCL9L | 5348 | 0.649 | 0.3699 | No | ||
| 82 | DYRK1A | 5380 | 0.637 | 0.3709 | No | ||
| 83 | PRDM2 | 5606 | 0.577 | 0.3611 | No | ||
| 84 | SMOC1 | 5690 | 0.552 | 0.3589 | No | ||
| 85 | EEF1E1 | 5768 | 0.532 | 0.3570 | No | ||
| 86 | THRAP2 | 5829 | 0.516 | 0.3559 | No | ||
| 87 | BAZ2A | 5872 | 0.508 | 0.3558 | No | ||
| 88 | RAB6IP1 | 5998 | 0.477 | 0.3510 | No | ||
| 89 | GRM1 | 6508 | 0.348 | 0.3248 | No | ||
| 90 | CORO1C | 6828 | 0.273 | 0.3087 | No | ||
| 91 | MAP1LC3B | 6929 | 0.247 | 0.3043 | No | ||
| 92 | SLC17A7 | 7047 | 0.222 | 0.2989 | No | ||
| 93 | NEUROG1 | 7204 | 0.188 | 0.2912 | No | ||
| 94 | SOX11 | 7214 | 0.185 | 0.2915 | No | ||
| 95 | PHF13 | 7399 | 0.142 | 0.2821 | No | ||
| 96 | ADAMTS9 | 7698 | 0.077 | 0.2663 | No | ||
| 97 | MRPL52 | 7889 | 0.033 | 0.2562 | No | ||
| 98 | SPRED1 | 7924 | 0.025 | 0.2544 | No | ||
| 99 | MRPL35 | 8141 | -0.025 | 0.2428 | No | ||
| 100 | ELL2 | 8163 | -0.031 | 0.2418 | No | ||
| 101 | SGCZ | 8308 | -0.062 | 0.2343 | No | ||
| 102 | FJX1 | 8541 | -0.114 | 0.2222 | No | ||
| 103 | ESRRG | 8806 | -0.163 | 0.2086 | No | ||
| 104 | EDG2 | 8993 | -0.206 | 0.1993 | No | ||
| 105 | ST7 | 9041 | -0.217 | 0.1977 | No | ||
| 106 | SLC22A2 | 9051 | -0.219 | 0.1981 | No | ||
| 107 | ESR1 | 9054 | -0.220 | 0.1989 | No | ||
| 108 | ELAVL3 | 9184 | -0.248 | 0.1930 | No | ||
| 109 | ZCCHC14 | 9521 | -0.324 | 0.1761 | No | ||
| 110 | SLITRK4 | 9527 | -0.325 | 0.1772 | No | ||
| 111 | SF3B1 | 9632 | -0.347 | 0.1730 | No | ||
| 112 | RPS6KC1 | 9774 | -0.375 | 0.1670 | No | ||
| 113 | PCDH9 | 10005 | -0.414 | 0.1562 | No | ||
| 114 | JPH3 | 10095 | -0.432 | 0.1532 | No | ||
| 115 | NPTX1 | 10441 | -0.496 | 0.1366 | No | ||
| 116 | ANGPT1 | 10667 | -0.538 | 0.1267 | No | ||
| 117 | KHDRBS3 | 10905 | -0.585 | 0.1163 | No | ||
| 118 | WNT4 | 10925 | -0.588 | 0.1177 | No | ||
| 119 | GABRB3 | 11208 | -0.644 | 0.1051 | No | ||
| 120 | CPNE8 | 11773 | -0.736 | 0.0776 | No | ||
| 121 | HIC2 | 11960 | -0.773 | 0.0708 | No | ||
| 122 | AP1S3 | 12004 | -0.782 | 0.0717 | No | ||
| 123 | GPM6A | 12345 | -0.842 | 0.0568 | No | ||
| 124 | DULLARD | 12656 | -0.899 | 0.0438 | No | ||
| 125 | HOXC8 | 12751 | -0.918 | 0.0425 | No | ||
| 126 | RHOBTB3 | 12872 | -0.940 | 0.0399 | No | ||
| 127 | BCL9 | 12875 | -0.940 | 0.0437 | No | ||
| 128 | P4HB | 13037 | -0.968 | 0.0391 | No | ||
| 129 | TTYH1 | 13406 | -1.039 | 0.0235 | No | ||
| 130 | EDEM1 | 13544 | -1.068 | 0.0205 | No | ||
| 131 | KCNA3 | 13684 | -1.092 | 0.0175 | No | ||
| 132 | CHN2 | 14076 | -1.167 | 0.0012 | No | ||
| 133 | SHC1 | 14136 | -1.182 | 0.0030 | No | ||
| 134 | AP2A2 | 14218 | -1.200 | 0.0036 | No | ||
| 135 | NR4A2 | 14288 | -1.217 | 0.0050 | No | ||
| 136 | DVL3 | 14330 | -1.228 | 0.0079 | No | ||
| 137 | ADCY6 | 14458 | -1.254 | 0.0062 | No | ||
| 138 | ING4 | 14547 | -1.275 | 0.0068 | No | ||
| 139 | SLC37A3 | 14655 | -1.295 | 0.0064 | No | ||
| 140 | MAP3K3 | 15290 | -1.438 | -0.0219 | No | ||
| 141 | EPHB6 | 15595 | -1.516 | -0.0321 | No | ||
| 142 | KCTD1 | 15600 | -1.517 | -0.0259 | No | ||
| 143 | SEC24D | 15629 | -1.525 | -0.0211 | No | ||
| 144 | TRPC5 | 15662 | -1.534 | -0.0164 | No | ||
| 145 | CDC25B | 15829 | -1.580 | -0.0188 | No | ||
| 146 | RSPO3 | 16027 | -1.639 | -0.0226 | No | ||
| 147 | FARP1 | 16039 | -1.642 | -0.0163 | No | ||
| 148 | FREM1 | 16070 | -1.649 | -0.0111 | No | ||
| 149 | CREB5 | 16342 | -1.724 | -0.0185 | No | ||
| 150 | AP1S1 | 16355 | -1.728 | -0.0120 | No | ||
| 151 | POU3F2 | 16562 | -1.806 | -0.0156 | No | ||
| 152 | ANXA11 | 16588 | -1.814 | -0.0094 | No | ||
| 153 | TGFBR2 | 16674 | -1.846 | -0.0062 | No | ||
| 154 | KLF12 | 16747 | -1.873 | -0.0023 | No | ||
| 155 | FBN2 | 17164 | -2.042 | -0.0163 | No | ||
| 156 | ELF2 | 17312 | -2.107 | -0.0155 | No | ||
| 157 | APH1A | 17539 | -2.238 | -0.0184 | No | ||
| 158 | GGA2 | 17896 | -2.492 | -0.0272 | No | ||
| 159 | DLG5 | 18030 | -2.628 | -0.0234 | No | ||
| 160 | DTX1 | 18212 | -2.848 | -0.0213 | No | ||
| 161 | CELSR3 | 18266 | -2.945 | -0.0119 | No | ||
| 162 | AUP1 | 18491 | -3.480 | -0.0095 | No | ||
| 163 | BCL2 | 18546 | -3.877 | 0.0038 | No |
