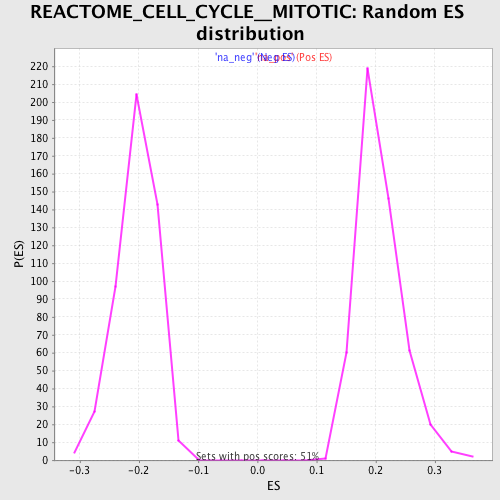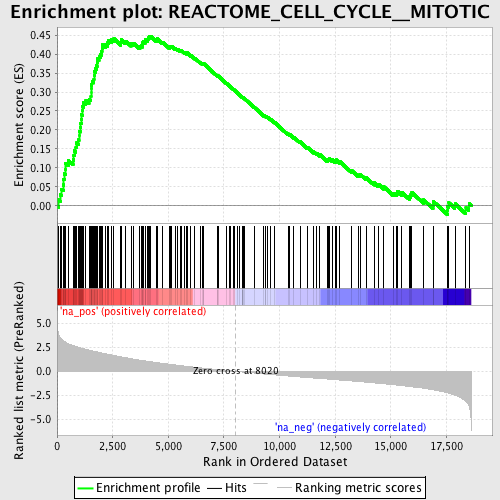
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.4472713 |
| Normalized Enrichment Score (NES) | 2.164295 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0041126376 |
| FWER p-Value | 0.019 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ORC4L | 68 | 3.893 | 0.0158 | Yes | ||
| 2 | NDC80 | 138 | 3.521 | 0.0298 | Yes | ||
| 3 | PSMD12 | 188 | 3.388 | 0.0441 | Yes | ||
| 4 | SMC1A | 277 | 3.191 | 0.0553 | Yes | ||
| 5 | CCNH | 296 | 3.158 | 0.0702 | Yes | ||
| 6 | PSMD14 | 320 | 3.126 | 0.0846 | Yes | ||
| 7 | MCM4 | 378 | 3.033 | 0.0968 | Yes | ||
| 8 | CCNA2 | 386 | 3.023 | 0.1115 | Yes | ||
| 9 | NUP107 | 499 | 2.875 | 0.1199 | Yes | ||
| 10 | MAD2L1 | 718 | 2.685 | 0.1215 | Yes | ||
| 11 | PSMA3 | 751 | 2.659 | 0.1331 | Yes | ||
| 12 | BUB1B | 774 | 2.637 | 0.1452 | Yes | ||
| 13 | PSMA2 | 846 | 2.581 | 0.1543 | Yes | ||
| 14 | PLK4 | 870 | 2.570 | 0.1659 | Yes | ||
| 15 | PSMA4 | 956 | 2.511 | 0.1739 | Yes | ||
| 16 | RFC1 | 985 | 2.492 | 0.1849 | Yes | ||
| 17 | FGFR1OP | 989 | 2.490 | 0.1972 | Yes | ||
| 18 | POLE | 1038 | 2.455 | 0.2069 | Yes | ||
| 19 | CCNB1 | 1062 | 2.442 | 0.2179 | Yes | ||
| 20 | PSMD7 | 1087 | 2.431 | 0.2288 | Yes | ||
| 21 | KIF23 | 1105 | 2.417 | 0.2400 | Yes | ||
| 22 | CDK2 | 1128 | 2.401 | 0.2509 | Yes | ||
| 23 | PSMD4 | 1139 | 2.395 | 0.2623 | Yes | ||
| 24 | OFD1 | 1185 | 2.367 | 0.2718 | Yes | ||
| 25 | RB1 | 1293 | 2.301 | 0.2775 | Yes | ||
| 26 | RPA1 | 1435 | 2.221 | 0.2810 | Yes | ||
| 27 | MAPRE1 | 1521 | 2.168 | 0.2873 | Yes | ||
| 28 | PCNA | 1525 | 2.167 | 0.2980 | Yes | ||
| 29 | RAD21 | 1527 | 2.167 | 0.3088 | Yes | ||
| 30 | PAFAH1B1 | 1532 | 2.165 | 0.3194 | Yes | ||
| 31 | RANBP2 | 1568 | 2.144 | 0.3283 | Yes | ||
| 32 | PRIM1 | 1654 | 2.096 | 0.3342 | Yes | ||
| 33 | NUP160 | 1660 | 2.095 | 0.3444 | Yes | ||
| 34 | SGOL2 | 1669 | 2.091 | 0.3545 | Yes | ||
| 35 | PSMD3 | 1737 | 2.057 | 0.3612 | Yes | ||
| 36 | CUL1 | 1780 | 2.035 | 0.3691 | Yes | ||
| 37 | ZW10 | 1798 | 2.028 | 0.3784 | Yes | ||
| 38 | NUP37 | 1819 | 2.018 | 0.3874 | Yes | ||
| 39 | YWHAG | 1884 | 1.980 | 0.3939 | Yes | ||
| 40 | POLD3 | 1952 | 1.938 | 0.3999 | Yes | ||
| 41 | CETN2 | 2005 | 1.917 | 0.4067 | Yes | ||
| 42 | SGOL1 | 2019 | 1.909 | 0.4156 | Yes | ||
| 43 | CDC45L | 2034 | 1.897 | 0.4244 | Yes | ||
| 44 | STAG2 | 2183 | 1.825 | 0.4255 | Yes | ||
| 45 | ORC5L | 2275 | 1.784 | 0.4295 | Yes | ||
| 46 | CDC25C | 2318 | 1.762 | 0.4361 | Yes | ||
| 47 | ZWINT | 2444 | 1.700 | 0.4378 | Yes | ||
| 48 | RPA3 | 2554 | 1.655 | 0.4402 | Yes | ||
| 49 | UBE2E1 | 2865 | 1.513 | 0.4310 | Yes | ||
| 50 | GMNN | 2881 | 1.506 | 0.4378 | Yes | ||
| 51 | KIF2A | 3083 | 1.417 | 0.4340 | Yes | ||
| 52 | MNAT1 | 3322 | 1.324 | 0.4278 | Yes | ||
| 53 | PSMC6 | 3436 | 1.279 | 0.4281 | Yes | ||
| 54 | RFC4 | 3704 | 1.173 | 0.4195 | Yes | ||
| 55 | KNTC1 | 3770 | 1.150 | 0.4217 | Yes | ||
| 56 | WEE1 | 3832 | 1.131 | 0.4241 | Yes | ||
| 57 | CDC16 | 3837 | 1.128 | 0.4295 | Yes | ||
| 58 | RANGAP1 | 3882 | 1.109 | 0.4327 | Yes | ||
| 59 | PSMA1 | 3953 | 1.082 | 0.4344 | Yes | ||
| 60 | PPP1CC | 3973 | 1.074 | 0.4387 | Yes | ||
| 61 | PSMD11 | 4075 | 1.037 | 0.4384 | Yes | ||
| 62 | ORC1L | 4093 | 1.032 | 0.4427 | Yes | ||
| 63 | PSMC5 | 4131 | 1.021 | 0.4458 | Yes | ||
| 64 | CCNB2 | 4198 | 1.002 | 0.4473 | Yes | ||
| 65 | BIRC5 | 4482 | 0.913 | 0.4365 | No | ||
| 66 | NUP43 | 4494 | 0.908 | 0.4405 | No | ||
| 67 | CENPA | 4741 | 0.829 | 0.4313 | No | ||
| 68 | PSME3 | 5056 | 0.736 | 0.4180 | No | ||
| 69 | PSMD2 | 5077 | 0.730 | 0.4206 | No | ||
| 70 | CLASP1 | 5155 | 0.707 | 0.4200 | No | ||
| 71 | ORC2L | 5301 | 0.665 | 0.4155 | No | ||
| 72 | FEN1 | 5409 | 0.630 | 0.4128 | No | ||
| 73 | SEC13 | 5532 | 0.598 | 0.4092 | No | ||
| 74 | CDC25A | 5598 | 0.579 | 0.4086 | No | ||
| 75 | DHFR | 5737 | 0.540 | 0.4039 | No | ||
| 76 | CDKN1A | 5813 | 0.522 | 0.4024 | No | ||
| 77 | YWHAE | 5843 | 0.513 | 0.4034 | No | ||
| 78 | CENPC1 | 6012 | 0.474 | 0.3967 | No | ||
| 79 | SDCCAG8 | 6195 | 0.425 | 0.3890 | No | ||
| 80 | TUBG1 | 6454 | 0.363 | 0.3768 | No | ||
| 81 | MCM10 | 6525 | 0.345 | 0.3748 | No | ||
| 82 | PSMB1 | 6577 | 0.334 | 0.3737 | No | ||
| 83 | PTTG1 | 6596 | 0.329 | 0.3744 | No | ||
| 84 | UBE2D1 | 7208 | 0.187 | 0.3422 | No | ||
| 85 | CLASP2 | 7217 | 0.184 | 0.3427 | No | ||
| 86 | DCTN3 | 7240 | 0.178 | 0.3424 | No | ||
| 87 | PSMC3 | 7619 | 0.093 | 0.3224 | No | ||
| 88 | XPO1 | 7732 | 0.068 | 0.3167 | No | ||
| 89 | PSMD8 | 7778 | 0.059 | 0.3146 | No | ||
| 90 | CCNA1 | 7930 | 0.025 | 0.3065 | No | ||
| 91 | UBE2C | 7954 | 0.017 | 0.3053 | No | ||
| 92 | DYNLL1 | 8124 | -0.022 | 0.2963 | No | ||
| 93 | RFC3 | 8179 | -0.034 | 0.2936 | No | ||
| 94 | PSMA6 | 8317 | -0.065 | 0.2865 | No | ||
| 95 | PSMB5 | 8324 | -0.067 | 0.2865 | No | ||
| 96 | PSMB9 | 8339 | -0.070 | 0.2861 | No | ||
| 97 | MCM3 | 8379 | -0.078 | 0.2844 | No | ||
| 98 | BUB1 | 8427 | -0.087 | 0.2822 | No | ||
| 99 | PSMB4 | 8875 | -0.181 | 0.2590 | No | ||
| 100 | POLA2 | 9292 | -0.275 | 0.2378 | No | ||
| 101 | E2F1 | 9361 | -0.288 | 0.2356 | No | ||
| 102 | MCM2 | 9380 | -0.291 | 0.2361 | No | ||
| 103 | TUBA4A | 9450 | -0.306 | 0.2339 | No | ||
| 104 | MCM5 | 9592 | -0.339 | 0.2279 | No | ||
| 105 | POLA1 | 9772 | -0.374 | 0.2201 | No | ||
| 106 | CLIP1 | 10384 | -0.487 | 0.1895 | No | ||
| 107 | PSMA5 | 10455 | -0.498 | 0.1882 | No | ||
| 108 | BUB3 | 10638 | -0.535 | 0.1810 | No | ||
| 109 | POLD2 | 10918 | -0.587 | 0.1689 | No | ||
| 110 | ACTR1A | 11242 | -0.650 | 0.1546 | No | ||
| 111 | NUMA1 | 11531 | -0.700 | 0.1426 | No | ||
| 112 | POLE2 | 11667 | -0.721 | 0.1389 | No | ||
| 113 | PSMC1 | 11815 | -0.743 | 0.1346 | No | ||
| 114 | CDK7 | 12156 | -0.809 | 0.1203 | No | ||
| 115 | SSNA1 | 12189 | -0.814 | 0.1226 | No | ||
| 116 | KIF20A | 12225 | -0.822 | 0.1249 | No | ||
| 117 | POLD1 | 12355 | -0.843 | 0.1221 | No | ||
| 118 | TYMS | 12508 | -0.872 | 0.1183 | No | ||
| 119 | PSMD5 | 12535 | -0.877 | 0.1212 | No | ||
| 120 | PSMA7 | 12700 | -0.906 | 0.1169 | No | ||
| 121 | PSMC4 | 13249 | -1.009 | 0.0923 | No | ||
| 122 | PRKAR2B | 13550 | -1.069 | 0.0814 | No | ||
| 123 | RFC5 | 13635 | -1.084 | 0.0823 | No | ||
| 124 | MCM7 | 13889 | -1.130 | 0.0743 | No | ||
| 125 | PRKACA | 14249 | -1.207 | 0.0609 | No | ||
| 126 | PSMB10 | 14455 | -1.254 | 0.0561 | No | ||
| 127 | CDC20 | 14677 | -1.299 | 0.0506 | No | ||
| 128 | LIG1 | 15125 | -1.400 | 0.0335 | No | ||
| 129 | PMF1 | 15252 | -1.430 | 0.0338 | No | ||
| 130 | PLK1 | 15307 | -1.442 | 0.0381 | No | ||
| 131 | CENPE | 15495 | -1.491 | 0.0355 | No | ||
| 132 | CDC25B | 15829 | -1.580 | 0.0254 | No | ||
| 133 | PSMB2 | 15902 | -1.601 | 0.0295 | No | ||
| 134 | NEK2 | 15945 | -1.615 | 0.0353 | No | ||
| 135 | CCNE1 | 16471 | -1.770 | 0.0158 | No | ||
| 136 | RPA2 | 16897 | -1.935 | 0.0025 | No | ||
| 137 | PSMD9 | 16925 | -1.948 | 0.0108 | No | ||
| 138 | DCTN2 | 17545 | -2.241 | -0.0115 | No | ||
| 139 | TUBA1A | 17553 | -2.244 | -0.0006 | No | ||
| 140 | TUBB4 | 17609 | -2.279 | 0.0078 | No | ||
| 141 | TK2 | 17884 | -2.480 | 0.0054 | No | ||
| 142 | CSNK1E | 18377 | -3.173 | -0.0053 | No | ||
| 143 | CDK4 | 18513 | -3.623 | 0.0056 | No |