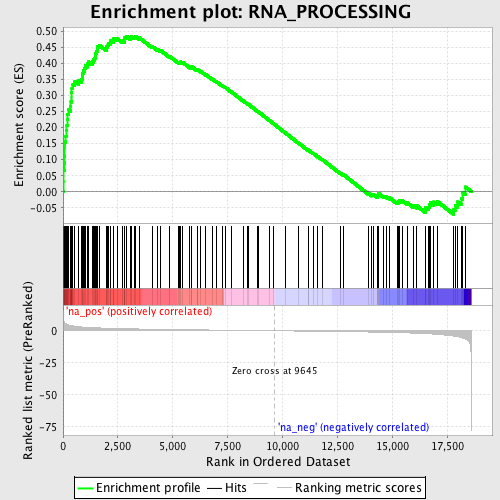
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | 0.48499298 |
| Normalized Enrichment Score (NES) | 2.111351 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0028088118 |
| FWER p-Value | 0.02 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FBL | 12 | 9.172 | 0.0336 | Yes | ||
| 2 | NOLA2 | 16 | 9.006 | 0.0670 | Yes | ||
| 3 | SNRPD3 | 51 | 6.620 | 0.0899 | Yes | ||
| 4 | NOLC1 | 74 | 6.118 | 0.1115 | Yes | ||
| 5 | PPAN | 82 | 5.990 | 0.1335 | Yes | ||
| 6 | U2AF1 | 86 | 5.951 | 0.1555 | Yes | ||
| 7 | SNRP70 | 124 | 5.420 | 0.1737 | Yes | ||
| 8 | PHF5A | 167 | 4.975 | 0.1900 | Yes | ||
| 9 | RRP9 | 174 | 4.897 | 0.2080 | Yes | ||
| 10 | SNRPA1 | 193 | 4.757 | 0.2248 | Yes | ||
| 11 | DDX39 | 205 | 4.706 | 0.2417 | Yes | ||
| 12 | EXOSC2 | 240 | 4.536 | 0.2568 | Yes | ||
| 13 | NSUN2 | 344 | 4.137 | 0.2667 | Yes | ||
| 14 | CRNKL1 | 357 | 4.084 | 0.2813 | Yes | ||
| 15 | LSM4 | 391 | 3.958 | 0.2942 | Yes | ||
| 16 | METTL1 | 392 | 3.956 | 0.3090 | Yes | ||
| 17 | CPSF1 | 402 | 3.914 | 0.3231 | Yes | ||
| 18 | SFRS5 | 446 | 3.781 | 0.3349 | Yes | ||
| 19 | AARS | 536 | 3.562 | 0.3434 | Yes | ||
| 20 | SNRPF | 691 | 3.192 | 0.3470 | Yes | ||
| 21 | SFRS9 | 832 | 2.959 | 0.3504 | Yes | ||
| 22 | TRNT1 | 869 | 2.895 | 0.3593 | Yes | ||
| 23 | SYNCRIP | 888 | 2.876 | 0.3691 | Yes | ||
| 24 | PABPC1 | 920 | 2.833 | 0.3779 | Yes | ||
| 25 | EFTUD2 | 987 | 2.757 | 0.3847 | Yes | ||
| 26 | POP4 | 1023 | 2.725 | 0.3929 | Yes | ||
| 27 | PABPC4 | 1103 | 2.640 | 0.3985 | Yes | ||
| 28 | SFRS2IP | 1168 | 2.583 | 0.4047 | Yes | ||
| 29 | CSTF3 | 1326 | 2.423 | 0.4052 | Yes | ||
| 30 | TXNL4A | 1374 | 2.384 | 0.4116 | Yes | ||
| 31 | RNMT | 1442 | 2.326 | 0.4167 | Yes | ||
| 32 | SFRS6 | 1463 | 2.309 | 0.4242 | Yes | ||
| 33 | NCBP2 | 1478 | 2.300 | 0.4320 | Yes | ||
| 34 | CDC2L2 | 1522 | 2.277 | 0.4382 | Yes | ||
| 35 | SF3A2 | 1560 | 2.246 | 0.4446 | Yes | ||
| 36 | EXOSC7 | 1579 | 2.234 | 0.4519 | Yes | ||
| 37 | GEMIN4 | 1641 | 2.198 | 0.4568 | Yes | ||
| 38 | SFRS2 | 1972 | 2.002 | 0.4465 | Yes | ||
| 39 | NOL5A | 1991 | 1.991 | 0.4529 | Yes | ||
| 40 | FUSIP1 | 2044 | 1.962 | 0.4574 | Yes | ||
| 41 | HNRPDL | 2082 | 1.943 | 0.4627 | Yes | ||
| 42 | CSTF2 | 2159 | 1.905 | 0.4657 | Yes | ||
| 43 | SIP1 | 2164 | 1.902 | 0.4726 | Yes | ||
| 44 | ZNF638 | 2300 | 1.839 | 0.4721 | Yes | ||
| 45 | UPF1 | 2312 | 1.833 | 0.4784 | Yes | ||
| 46 | DDX20 | 2471 | 1.756 | 0.4764 | Yes | ||
| 47 | PRPF31 | 2704 | 1.645 | 0.4700 | Yes | ||
| 48 | KIN | 2788 | 1.617 | 0.4715 | Yes | ||
| 49 | SF3A3 | 2790 | 1.616 | 0.4775 | Yes | ||
| 50 | PRPF3 | 2815 | 1.605 | 0.4822 | Yes | ||
| 51 | RBM6 | 2877 | 1.586 | 0.4848 | Yes | ||
| 52 | DBR1 | 3060 | 1.519 | 0.4806 | Yes | ||
| 53 | ELAVL4 | 3125 | 1.496 | 0.4827 | Yes | ||
| 54 | EXOSC3 | 3235 | 1.457 | 0.4823 | Yes | ||
| 55 | CSTF1 | 3285 | 1.437 | 0.4850 | Yes | ||
| 56 | DGCR8 | 3475 | 1.372 | 0.4799 | No | ||
| 57 | IVNS1ABP | 4052 | 1.199 | 0.4532 | No | ||
| 58 | GEMIN5 | 4289 | 1.137 | 0.4447 | No | ||
| 59 | SNRPD2 | 4445 | 1.100 | 0.4404 | No | ||
| 60 | SLBP | 4852 | 1.001 | 0.4222 | No | ||
| 61 | SFRS10 | 5277 | 0.897 | 0.4026 | No | ||
| 62 | PTBP1 | 5324 | 0.885 | 0.4034 | No | ||
| 63 | GRSF1 | 5364 | 0.873 | 0.4046 | No | ||
| 64 | SNRPB | 5454 | 0.853 | 0.4030 | No | ||
| 65 | PPARGC1A | 5777 | 0.779 | 0.3885 | No | ||
| 66 | SNRPD1 | 5854 | 0.764 | 0.3872 | No | ||
| 67 | RBMY1A1 | 5868 | 0.760 | 0.3893 | No | ||
| 68 | SSB | 6108 | 0.708 | 0.3791 | No | ||
| 69 | SF3B4 | 6132 | 0.705 | 0.3804 | No | ||
| 70 | NOL3 | 6276 | 0.673 | 0.3752 | No | ||
| 71 | DHX15 | 6496 | 0.629 | 0.3657 | No | ||
| 72 | GEMIN7 | 6800 | 0.569 | 0.3515 | No | ||
| 73 | HNRPD | 6999 | 0.530 | 0.3427 | No | ||
| 74 | SF3B3 | 7261 | 0.473 | 0.3304 | No | ||
| 75 | FARS2 | 7385 | 0.447 | 0.3254 | No | ||
| 76 | LSM3 | 7676 | 0.392 | 0.3112 | No | ||
| 77 | SNW1 | 8204 | 0.280 | 0.2837 | No | ||
| 78 | SF3A1 | 8407 | 0.245 | 0.2737 | No | ||
| 79 | HNRPF | 8445 | 0.236 | 0.2726 | No | ||
| 80 | RBPMS | 8867 | 0.151 | 0.2504 | No | ||
| 81 | NUFIP1 | 8893 | 0.147 | 0.2496 | No | ||
| 82 | BCAS2 | 9420 | 0.045 | 0.2213 | No | ||
| 83 | SNRPG | 9580 | 0.011 | 0.2128 | No | ||
| 84 | RNPS1 | 10151 | -0.102 | 0.1823 | No | ||
| 85 | SRPK2 | 10738 | -0.222 | 0.1515 | No | ||
| 86 | SFRS1 | 11161 | -0.311 | 0.1298 | No | ||
| 87 | DHX16 | 11171 | -0.313 | 0.1305 | No | ||
| 88 | NUDT21 | 11406 | -0.363 | 0.1192 | No | ||
| 89 | LSM5 | 11610 | -0.407 | 0.1098 | No | ||
| 90 | DHX38 | 11803 | -0.453 | 0.1011 | No | ||
| 91 | RBMS1 | 12639 | -0.637 | 0.0583 | No | ||
| 92 | CPSF3 | 12779 | -0.668 | 0.0533 | No | ||
| 93 | KHDRBS1 | 13897 | -0.956 | -0.0036 | No | ||
| 94 | NONO | 14077 | -1.006 | -0.0095 | No | ||
| 95 | USP39 | 14135 | -1.023 | -0.0087 | No | ||
| 96 | INTS6 | 14318 | -1.080 | -0.0146 | No | ||
| 97 | SNRPB2 | 14323 | -1.082 | -0.0107 | No | ||
| 98 | SPOP | 14355 | -1.090 | -0.0083 | No | ||
| 99 | ADAT1 | 14379 | -1.097 | -0.0055 | No | ||
| 100 | ASCC3L1 | 14615 | -1.169 | -0.0138 | No | ||
| 101 | RPS14 | 14720 | -1.203 | -0.0150 | No | ||
| 102 | PRPF4B | 14864 | -1.262 | -0.0180 | No | ||
| 103 | SF3B2 | 15217 | -1.408 | -0.0318 | No | ||
| 104 | SFPQ | 15284 | -1.436 | -0.0300 | No | ||
| 105 | PRPF8 | 15333 | -1.458 | -0.0271 | No | ||
| 106 | RNGTT | 15450 | -1.526 | -0.0277 | No | ||
| 107 | SFRS7 | 15673 | -1.651 | -0.0336 | No | ||
| 108 | IGF2BP3 | 15987 | -1.838 | -0.0436 | No | ||
| 109 | BICD1 | 16118 | -1.923 | -0.0435 | No | ||
| 110 | DUSP11 | 16512 | -2.224 | -0.0564 | No | ||
| 111 | DDX54 | 16528 | -2.236 | -0.0489 | No | ||
| 112 | GEMIN6 | 16672 | -2.364 | -0.0478 | No | ||
| 113 | RBM3 | 16708 | -2.401 | -0.0407 | No | ||
| 114 | RBM5 | 16752 | -2.458 | -0.0339 | No | ||
| 115 | SRRM1 | 16893 | -2.602 | -0.0318 | No | ||
| 116 | DHX8 | 17058 | -2.800 | -0.0302 | No | ||
| 117 | SFRS8 | 17809 | -4.128 | -0.0553 | No | ||
| 118 | SRPK1 | 17868 | -4.293 | -0.0424 | No | ||
| 119 | HNRPUL1 | 17992 | -4.683 | -0.0316 | No | ||
| 120 | ZNF346 | 18169 | -5.435 | -0.0209 | No | ||
| 121 | CUGBP1 | 18217 | -5.616 | -0.0024 | No | ||
| 122 | SNRPN | 18339 | -6.422 | 0.0150 | No |
