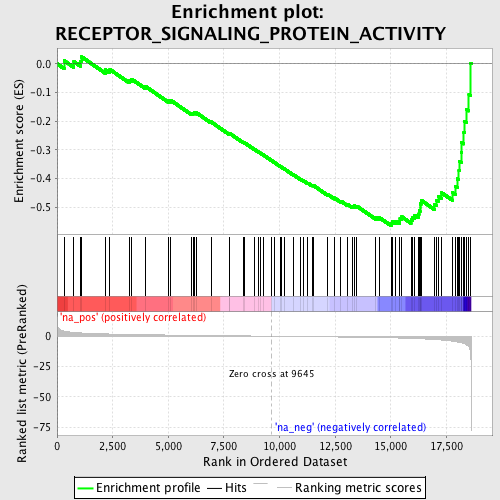
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RECEPTOR_SIGNALING_PROTEIN_ACTIVITY |
| Enrichment Score (ES) | -0.56601644 |
| Normalized Enrichment Score (NES) | -1.8586555 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.6172988 |
| FWER p-Value | 0.859 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TBL3 | 318 | 4.247 | 0.0108 | No | ||
| 2 | IL1F5 | 753 | 3.086 | 0.0078 | No | ||
| 3 | SP110 | 1049 | 2.704 | 0.0097 | No | ||
| 4 | IL1RL1 | 1102 | 2.641 | 0.0243 | No | ||
| 5 | LYN | 2171 | 1.899 | -0.0208 | No | ||
| 6 | IFITM1 | 2372 | 1.805 | -0.0197 | No | ||
| 7 | BAG1 | 3244 | 1.454 | -0.0571 | No | ||
| 8 | TYRO3 | 3347 | 1.413 | -0.0533 | No | ||
| 9 | MAP3K6 | 3960 | 1.225 | -0.0782 | No | ||
| 10 | MAP3K7 | 5000 | 0.970 | -0.1278 | No | ||
| 11 | MAPK10 | 5114 | 0.942 | -0.1277 | No | ||
| 12 | NCR1 | 6050 | 0.720 | -0.1734 | No | ||
| 13 | IRAK1 | 6113 | 0.708 | -0.1721 | No | ||
| 14 | MAPK13 | 6168 | 0.698 | -0.1704 | No | ||
| 15 | PPARGC1B | 6262 | 0.675 | -0.1710 | No | ||
| 16 | MAP3K9 | 6919 | 0.544 | -0.2027 | No | ||
| 17 | SMAD7 | 7752 | 0.378 | -0.2451 | No | ||
| 18 | CSPG4 | 7757 | 0.377 | -0.2429 | No | ||
| 19 | ACVR2A | 8372 | 0.250 | -0.2743 | No | ||
| 20 | ACVR1B | 8440 | 0.237 | -0.2764 | No | ||
| 21 | ACVR1C | 8891 | 0.147 | -0.2997 | No | ||
| 22 | MAP3K10 | 9073 | 0.112 | -0.3087 | No | ||
| 23 | GNAZ | 9137 | 0.101 | -0.3114 | No | ||
| 24 | ARF1 | 9284 | 0.074 | -0.3188 | No | ||
| 25 | ACVR1 | 9653 | -0.003 | -0.3386 | No | ||
| 26 | SKIL | 9772 | -0.022 | -0.3449 | No | ||
| 27 | MAPK4 | 10018 | -0.073 | -0.3576 | No | ||
| 28 | ACVRL1 | 10103 | -0.091 | -0.3615 | No | ||
| 29 | MAP3K13 | 10237 | -0.118 | -0.3679 | No | ||
| 30 | IL1RN | 10619 | -0.195 | -0.3872 | No | ||
| 31 | CD19 | 10950 | -0.264 | -0.4032 | No | ||
| 32 | NSMAF | 11082 | -0.295 | -0.4083 | No | ||
| 33 | TYROBP | 11274 | -0.333 | -0.4164 | No | ||
| 34 | MAP3K4 | 11463 | -0.376 | -0.4241 | No | ||
| 35 | SMAD6 | 11501 | -0.385 | -0.4236 | No | ||
| 36 | ERBB2 | 12170 | -0.526 | -0.4561 | No | ||
| 37 | BLNK | 12466 | -0.598 | -0.4681 | No | ||
| 38 | MAPK7 | 12748 | -0.663 | -0.4789 | No | ||
| 39 | KIT | 13043 | -0.731 | -0.4899 | No | ||
| 40 | BAG4 | 13261 | -0.790 | -0.4964 | No | ||
| 41 | IRS1 | 13344 | -0.810 | -0.4955 | No | ||
| 42 | CCL4 | 13467 | -0.840 | -0.4965 | No | ||
| 43 | ALK | 14320 | -1.081 | -0.5354 | No | ||
| 44 | TGFBR1 | 14475 | -1.126 | -0.5362 | No | ||
| 45 | MAPK8 | 15028 | -1.330 | -0.5573 | Yes | ||
| 46 | IL18BP | 15054 | -1.340 | -0.5498 | Yes | ||
| 47 | IRAK2 | 15218 | -1.408 | -0.5493 | Yes | ||
| 48 | MAPK14 | 15392 | -1.490 | -0.5488 | Yes | ||
| 49 | MAPK12 | 15410 | -1.504 | -0.5398 | Yes | ||
| 50 | MAP4K1 | 15494 | -1.551 | -0.5340 | Yes | ||
| 51 | MAP3K5 | 15909 | -1.779 | -0.5446 | Yes | ||
| 52 | MAPK1 | 15982 | -1.834 | -0.5364 | Yes | ||
| 53 | NLK | 16066 | -1.886 | -0.5285 | Yes | ||
| 54 | AMHR2 | 16232 | -2.005 | -0.5242 | Yes | ||
| 55 | INSR | 16269 | -2.034 | -0.5127 | Yes | ||
| 56 | PLCE1 | 16317 | -2.076 | -0.5016 | Yes | ||
| 57 | SHC1 | 16319 | -2.078 | -0.4879 | Yes | ||
| 58 | ACVR2B | 16359 | -2.103 | -0.4762 | Yes | ||
| 59 | MAP3K11 | 16964 | -2.684 | -0.4911 | Yes | ||
| 60 | DAXX | 17045 | -2.781 | -0.4770 | Yes | ||
| 61 | MAPK9 | 17129 | -2.896 | -0.4624 | Yes | ||
| 62 | ARNT | 17271 | -3.107 | -0.4496 | Yes | ||
| 63 | CDK5 | 17776 | -4.065 | -0.4500 | Yes | ||
| 64 | MAPK11 | 17912 | -4.438 | -0.4280 | Yes | ||
| 65 | STAT3 | 17998 | -4.700 | -0.4016 | Yes | ||
| 66 | MAP3K3 | 18061 | -4.882 | -0.3728 | Yes | ||
| 67 | SOCS2 | 18077 | -4.952 | -0.3409 | Yes | ||
| 68 | STAT2 | 18166 | -5.430 | -0.3099 | Yes | ||
| 69 | IKBKE | 18185 | -5.485 | -0.2747 | Yes | ||
| 70 | CD3E | 18282 | -6.048 | -0.2400 | Yes | ||
| 71 | TGFBR2 | 18307 | -6.182 | -0.2006 | Yes | ||
| 72 | STAT1 | 18408 | -7.085 | -0.1593 | Yes | ||
| 73 | CDKN1B | 18510 | -8.697 | -0.1074 | Yes | ||
| 74 | IL4R | 18594 | -17.159 | 0.0012 | Yes |
