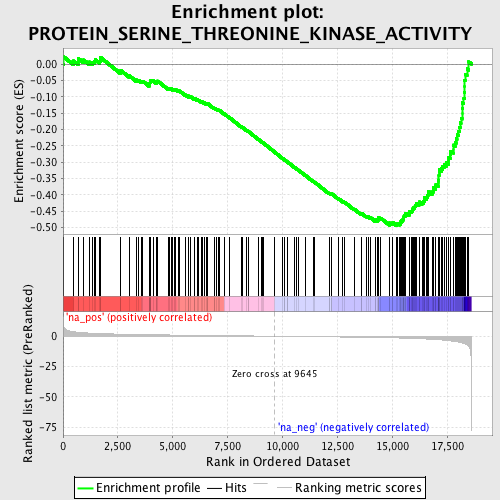
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY |
| Enrichment Score (ES) | -0.4950432 |
| Normalized Enrichment Score (NES) | -1.8393114 |
| Nominal p-value | 0.0011947431 |
| FDR q-value | 0.44022653 |
| FWER p-Value | 0.896 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RIPK4 | 28 | 7.789 | 0.0222 | No | ||
| 2 | MAPKAPK2 | 468 | 3.735 | 0.0098 | No | ||
| 3 | RIPK1 | 687 | 3.202 | 0.0077 | No | ||
| 4 | LATS2 | 698 | 3.175 | 0.0169 | No | ||
| 5 | NEK4 | 933 | 2.820 | 0.0128 | No | ||
| 6 | RPS6KA1 | 1191 | 2.561 | 0.0066 | No | ||
| 7 | EIF2AK2 | 1341 | 2.409 | 0.0059 | No | ||
| 8 | CDC42BPB | 1418 | 2.342 | 0.0089 | No | ||
| 9 | CHEK2 | 1465 | 2.308 | 0.0135 | No | ||
| 10 | RPS6KA3 | 1667 | 2.178 | 0.0092 | No | ||
| 11 | STK3 | 1699 | 2.159 | 0.0141 | No | ||
| 12 | ROCK2 | 1708 | 2.154 | 0.0202 | No | ||
| 13 | PRKAG3 | 2626 | 1.683 | -0.0244 | No | ||
| 14 | CDK9 | 2630 | 1.680 | -0.0194 | No | ||
| 15 | RPS6KA5 | 3014 | 1.536 | -0.0355 | No | ||
| 16 | ERN1 | 3331 | 1.420 | -0.0483 | No | ||
| 17 | TTK | 3434 | 1.386 | -0.0496 | No | ||
| 18 | RIPK3 | 3560 | 1.346 | -0.0523 | No | ||
| 19 | CSNK1D | 3626 | 1.327 | -0.0518 | No | ||
| 20 | RPS6KC1 | 3917 | 1.236 | -0.0637 | No | ||
| 21 | PLK4 | 3925 | 1.235 | -0.0603 | No | ||
| 22 | GTF2H1 | 3951 | 1.227 | -0.0579 | No | ||
| 23 | MAP3K6 | 3960 | 1.225 | -0.0546 | No | ||
| 24 | TTN | 3978 | 1.220 | -0.0519 | No | ||
| 25 | CSNK2B | 3994 | 1.216 | -0.0490 | No | ||
| 26 | STK19 | 4099 | 1.187 | -0.0510 | No | ||
| 27 | CIT | 4246 | 1.147 | -0.0554 | No | ||
| 28 | TESK1 | 4248 | 1.146 | -0.0520 | No | ||
| 29 | BCR | 4306 | 1.134 | -0.0516 | No | ||
| 30 | MARK1 | 4790 | 1.017 | -0.0747 | No | ||
| 31 | RPS6KA2 | 4865 | 0.998 | -0.0757 | No | ||
| 32 | DAPK3 | 4918 | 0.988 | -0.0755 | No | ||
| 33 | MAP3K7 | 5000 | 0.970 | -0.0769 | No | ||
| 34 | STK38 | 5096 | 0.947 | -0.0792 | No | ||
| 35 | MAPK10 | 5114 | 0.942 | -0.0772 | No | ||
| 36 | IRAK3 | 5241 | 0.908 | -0.0813 | No | ||
| 37 | MAPKAPK5 | 5303 | 0.891 | -0.0819 | No | ||
| 38 | TSSK2 | 5577 | 0.826 | -0.0942 | No | ||
| 39 | TESK2 | 5709 | 0.797 | -0.0988 | No | ||
| 40 | HUNK | 5726 | 0.792 | -0.0973 | No | ||
| 41 | EXOSC10 | 5824 | 0.771 | -0.1002 | No | ||
| 42 | BCKDK | 5975 | 0.738 | -0.1061 | No | ||
| 43 | PLK1 | 6008 | 0.731 | -0.1056 | No | ||
| 44 | IRAK1 | 6113 | 0.708 | -0.1091 | No | ||
| 45 | MAPK13 | 6168 | 0.698 | -0.1099 | No | ||
| 46 | BRSK2 | 6299 | 0.669 | -0.1149 | No | ||
| 47 | ROCK1 | 6347 | 0.656 | -0.1154 | No | ||
| 48 | PRKAG1 | 6428 | 0.642 | -0.1178 | No | ||
| 49 | CSNK2A2 | 6537 | 0.621 | -0.1218 | No | ||
| 50 | RPS6KB2 | 6565 | 0.616 | -0.1214 | No | ||
| 51 | VRK2 | 6598 | 0.609 | -0.1212 | No | ||
| 52 | AKT1 | 6886 | 0.551 | -0.1351 | No | ||
| 53 | MAP3K9 | 6919 | 0.544 | -0.1352 | No | ||
| 54 | LIMK1 | 6995 | 0.530 | -0.1377 | No | ||
| 55 | PHKG2 | 7072 | 0.517 | -0.1402 | No | ||
| 56 | MAST1 | 7115 | 0.508 | -0.1409 | No | ||
| 57 | NEK2 | 7349 | 0.455 | -0.1522 | No | ||
| 58 | CDK7 | 7589 | 0.409 | -0.1639 | No | ||
| 59 | TSSK6 | 8108 | 0.301 | -0.1911 | No | ||
| 60 | DAPK2 | 8178 | 0.287 | -0.1939 | No | ||
| 61 | ACVR2A | 8372 | 0.250 | -0.2036 | No | ||
| 62 | ACVR1B | 8440 | 0.237 | -0.2065 | No | ||
| 63 | PRKCZ | 8446 | 0.236 | -0.2061 | No | ||
| 64 | ACVR1C | 8891 | 0.147 | -0.2297 | No | ||
| 65 | TLK1 | 9043 | 0.117 | -0.2375 | No | ||
| 66 | MAP3K10 | 9073 | 0.112 | -0.2388 | No | ||
| 67 | CHUK | 9136 | 0.102 | -0.2418 | No | ||
| 68 | ACVR1 | 9653 | -0.003 | -0.2698 | No | ||
| 69 | MAPK4 | 10018 | -0.073 | -0.2893 | No | ||
| 70 | WNK3 | 10069 | -0.085 | -0.2918 | No | ||
| 71 | ACVRL1 | 10103 | -0.091 | -0.2933 | No | ||
| 72 | MAP3K13 | 10237 | -0.118 | -0.3001 | No | ||
| 73 | TAOK2 | 10248 | -0.120 | -0.3003 | No | ||
| 74 | ERN2 | 10560 | -0.181 | -0.3166 | No | ||
| 75 | SGK2 | 10617 | -0.195 | -0.3191 | No | ||
| 76 | SRPK2 | 10738 | -0.222 | -0.3249 | No | ||
| 77 | TSSK3 | 11026 | -0.283 | -0.3396 | No | ||
| 78 | VRK1 | 11409 | -0.363 | -0.3592 | No | ||
| 79 | MAP3K4 | 11463 | -0.376 | -0.3609 | No | ||
| 80 | MKNK2 | 12122 | -0.517 | -0.3950 | No | ||
| 81 | PRKCE | 12144 | -0.521 | -0.3946 | No | ||
| 82 | EIF2AK4 | 12209 | -0.535 | -0.3964 | No | ||
| 83 | PBK | 12217 | -0.537 | -0.3952 | No | ||
| 84 | CSNK2A1 | 12552 | -0.617 | -0.4114 | No | ||
| 85 | MAPK7 | 12748 | -0.663 | -0.4199 | No | ||
| 86 | STK16 | 12834 | -0.682 | -0.4225 | No | ||
| 87 | PRKCI | 13267 | -0.791 | -0.4435 | No | ||
| 88 | PAK2 | 13592 | -0.871 | -0.4584 | No | ||
| 89 | NEK11 | 13594 | -0.872 | -0.4558 | No | ||
| 90 | CDC42BPA | 13817 | -0.932 | -0.4650 | No | ||
| 91 | MINK1 | 13903 | -0.959 | -0.4667 | No | ||
| 92 | EIF2AK1 | 13989 | -0.984 | -0.4683 | No | ||
| 93 | STK38L | 14219 | -1.051 | -0.4775 | No | ||
| 94 | PRKACB | 14256 | -1.060 | -0.4762 | No | ||
| 95 | CDKL1 | 14336 | -1.085 | -0.4772 | No | ||
| 96 | TSSK1A | 14358 | -1.091 | -0.4750 | No | ||
| 97 | MAP3K2 | 14359 | -1.091 | -0.4717 | No | ||
| 98 | MAPK6 | 14362 | -1.092 | -0.4685 | No | ||
| 99 | TGFBR1 | 14475 | -1.126 | -0.4711 | No | ||
| 100 | PAK1 | 14871 | -1.265 | -0.4887 | No | ||
| 101 | ILK | 14888 | -1.271 | -0.4857 | No | ||
| 102 | MARK4 | 15005 | -1.319 | -0.4879 | No | ||
| 103 | MAPK8 | 15028 | -1.330 | -0.4851 | No | ||
| 104 | WNK4 | 15213 | -1.406 | -0.4908 | Yes | ||
| 105 | IRAK2 | 15218 | -1.408 | -0.4867 | Yes | ||
| 106 | HIPK3 | 15322 | -1.453 | -0.4878 | Yes | ||
| 107 | CDK3 | 15389 | -1.489 | -0.4869 | Yes | ||
| 108 | MAPK14 | 15392 | -1.490 | -0.4825 | Yes | ||
| 109 | MAPK12 | 15410 | -1.504 | -0.4788 | Yes | ||
| 110 | ATM | 15455 | -1.529 | -0.4765 | Yes | ||
| 111 | MAP4K1 | 15494 | -1.551 | -0.4738 | Yes | ||
| 112 | PHKA1 | 15506 | -1.559 | -0.4697 | Yes | ||
| 113 | CPNE3 | 15533 | -1.574 | -0.4663 | Yes | ||
| 114 | CDK10 | 15545 | -1.583 | -0.4621 | Yes | ||
| 115 | CLK1 | 15599 | -1.608 | -0.4601 | Yes | ||
| 116 | PRKCB1 | 15618 | -1.616 | -0.4561 | Yes | ||
| 117 | STK17B | 15768 | -1.700 | -0.4590 | Yes | ||
| 118 | UHMK1 | 15785 | -1.710 | -0.4547 | Yes | ||
| 119 | MYLK | 15792 | -1.714 | -0.4498 | Yes | ||
| 120 | RIPK2 | 15887 | -1.764 | -0.4495 | Yes | ||
| 121 | MAP3K5 | 15909 | -1.779 | -0.4452 | Yes | ||
| 122 | PHKA2 | 15944 | -1.805 | -0.4415 | Yes | ||
| 123 | MAPK1 | 15982 | -1.834 | -0.4380 | Yes | ||
| 124 | STK11 | 16037 | -1.869 | -0.4352 | Yes | ||
| 125 | NLK | 16066 | -1.886 | -0.4310 | Yes | ||
| 126 | CSNK1A1 | 16088 | -1.901 | -0.4263 | Yes | ||
| 127 | NEK6 | 16221 | -1.999 | -0.4274 | Yes | ||
| 128 | MAP4K5 | 16226 | -2.001 | -0.4215 | Yes | ||
| 129 | ACVR2B | 16359 | -2.103 | -0.4222 | Yes | ||
| 130 | PRKCH | 16424 | -2.159 | -0.4191 | Yes | ||
| 131 | PRKD1 | 16464 | -2.190 | -0.4146 | Yes | ||
| 132 | MKNK1 | 16480 | -2.203 | -0.4087 | Yes | ||
| 133 | AKT2 | 16584 | -2.289 | -0.4073 | Yes | ||
| 134 | PLK3 | 16606 | -2.308 | -0.4014 | Yes | ||
| 135 | LIMK2 | 16635 | -2.331 | -0.3958 | Yes | ||
| 136 | MAP4K4 | 16650 | -2.348 | -0.3894 | Yes | ||
| 137 | PINK1 | 16818 | -2.518 | -0.3908 | Yes | ||
| 138 | LMTK2 | 16862 | -2.568 | -0.3853 | Yes | ||
| 139 | STK4 | 16872 | -2.580 | -0.3779 | Yes | ||
| 140 | MAP3K11 | 16964 | -2.684 | -0.3747 | Yes | ||
| 141 | PRKD3 | 16992 | -2.712 | -0.3679 | Yes | ||
| 142 | CDK2 | 17094 | -2.849 | -0.3647 | Yes | ||
| 143 | WNK1 | 17096 | -2.850 | -0.3560 | Yes | ||
| 144 | PRKX | 17128 | -2.891 | -0.3489 | Yes | ||
| 145 | MAPK9 | 17129 | -2.896 | -0.3401 | Yes | ||
| 146 | GSK3B | 17139 | -2.906 | -0.3317 | Yes | ||
| 147 | TLK2 | 17145 | -2.916 | -0.3231 | Yes | ||
| 148 | STK10 | 17242 | -3.070 | -0.3189 | Yes | ||
| 149 | CSNK1G2 | 17295 | -3.137 | -0.3122 | Yes | ||
| 150 | ULK1 | 17368 | -3.262 | -0.3062 | Yes | ||
| 151 | PIM2 | 17476 | -3.462 | -0.3014 | Yes | ||
| 152 | MAP4K3 | 17566 | -3.626 | -0.2952 | Yes | ||
| 153 | EIF2AK3 | 17575 | -3.644 | -0.2845 | Yes | ||
| 154 | MAP3K8 | 17644 | -3.789 | -0.2767 | Yes | ||
| 155 | CLK2 | 17661 | -3.812 | -0.2659 | Yes | ||
| 156 | CDK5 | 17776 | -4.065 | -0.2597 | Yes | ||
| 157 | MAP3K12 | 17778 | -4.065 | -0.2474 | Yes | ||
| 158 | SRPK1 | 17868 | -4.293 | -0.2391 | Yes | ||
| 159 | MAPK11 | 17912 | -4.438 | -0.2279 | Yes | ||
| 160 | SNF1LK | 17954 | -4.564 | -0.2162 | Yes | ||
| 161 | SNF1LK2 | 18018 | -4.752 | -0.2052 | Yes | ||
| 162 | MAP3K3 | 18061 | -4.882 | -0.1926 | Yes | ||
| 163 | PIM1 | 18097 | -5.045 | -0.1791 | Yes | ||
| 164 | BRDT | 18136 | -5.200 | -0.1653 | Yes | ||
| 165 | IKBKE | 18185 | -5.485 | -0.1512 | Yes | ||
| 166 | BRD2 | 18187 | -5.490 | -0.1345 | Yes | ||
| 167 | MARK2 | 18200 | -5.546 | -0.1183 | Yes | ||
| 168 | PDPK1 | 18265 | -5.919 | -0.1037 | Yes | ||
| 169 | CSNK1E | 18290 | -6.103 | -0.0864 | Yes | ||
| 170 | DYRK1A | 18303 | -6.167 | -0.0682 | Yes | ||
| 171 | TGFBR2 | 18307 | -6.182 | -0.0496 | Yes | ||
| 172 | SNRK | 18324 | -6.276 | -0.0313 | Yes | ||
| 173 | MAP4K2 | 18412 | -7.157 | -0.0142 | Yes | ||
| 174 | SGK3 | 18490 | -8.275 | 0.0068 | Yes |
