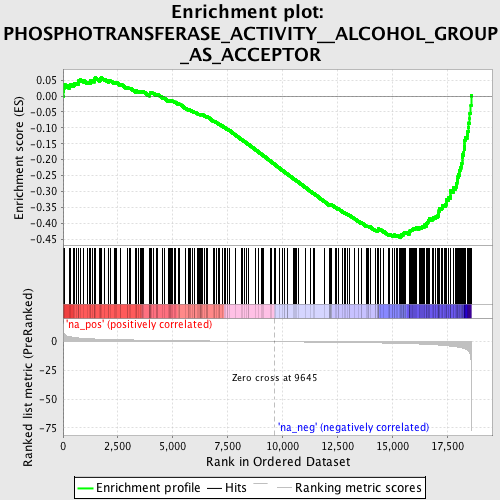
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PHOSPHOTRANSFERASE_ACTIVITY__ALCOHOL_GROUP_AS_ACCEPTOR |
| Enrichment Score (ES) | -0.44509703 |
| Normalized Enrichment Score (NES) | -1.7402791 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.31757346 |
| FWER p-Value | 0.995 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PKM2 | 9 | 9.424 | 0.0156 | No | ||
| 2 | RIPK4 | 28 | 7.789 | 0.0279 | No | ||
| 3 | PFKM | 79 | 6.059 | 0.0356 | No | ||
| 4 | PIK3R3 | 276 | 4.399 | 0.0324 | No | ||
| 5 | GALK1 | 326 | 4.217 | 0.0369 | No | ||
| 6 | MAPKAPK2 | 468 | 3.735 | 0.0356 | No | ||
| 7 | MAP3K7IP1 | 524 | 3.584 | 0.0387 | No | ||
| 8 | PFKL | 595 | 3.417 | 0.0407 | No | ||
| 9 | RIPK1 | 687 | 3.202 | 0.0412 | No | ||
| 10 | LATS2 | 698 | 3.175 | 0.0461 | No | ||
| 11 | PDGFRB | 718 | 3.142 | 0.0505 | No | ||
| 12 | EPHB2 | 786 | 3.039 | 0.0520 | No | ||
| 13 | NEK4 | 933 | 2.820 | 0.0488 | No | ||
| 14 | MVK | 1123 | 2.621 | 0.0430 | No | ||
| 15 | RPS6KA1 | 1191 | 2.561 | 0.0437 | No | ||
| 16 | FLT4 | 1254 | 2.501 | 0.0446 | No | ||
| 17 | DDR2 | 1255 | 2.500 | 0.0489 | No | ||
| 18 | EIF2AK2 | 1341 | 2.409 | 0.0484 | No | ||
| 19 | CDC42BPB | 1418 | 2.342 | 0.0482 | No | ||
| 20 | PIK3R2 | 1444 | 2.324 | 0.0508 | No | ||
| 21 | TGFA | 1447 | 2.323 | 0.0547 | No | ||
| 22 | CHEK2 | 1465 | 2.308 | 0.0577 | No | ||
| 23 | RPS6KA3 | 1667 | 2.178 | 0.0504 | No | ||
| 24 | STK3 | 1699 | 2.159 | 0.0524 | No | ||
| 25 | ROCK2 | 1708 | 2.154 | 0.0557 | No | ||
| 26 | HK2 | 1730 | 2.140 | 0.0582 | No | ||
| 27 | EPGN | 1887 | 2.045 | 0.0532 | No | ||
| 28 | PDK4 | 2076 | 1.949 | 0.0463 | No | ||
| 29 | TYK2 | 2086 | 1.942 | 0.0491 | No | ||
| 30 | LYN | 2171 | 1.899 | 0.0477 | No | ||
| 31 | DDR1 | 2333 | 1.823 | 0.0421 | No | ||
| 32 | ITPKA | 2381 | 1.803 | 0.0426 | No | ||
| 33 | FLT1 | 2437 | 1.773 | 0.0426 | No | ||
| 34 | PRKAG3 | 2626 | 1.683 | 0.0352 | No | ||
| 35 | CDK9 | 2630 | 1.680 | 0.0379 | No | ||
| 36 | GNE | 2916 | 1.571 | 0.0251 | No | ||
| 37 | RYK | 2944 | 1.559 | 0.0263 | No | ||
| 38 | RPS6KA5 | 3014 | 1.536 | 0.0251 | No | ||
| 39 | ROR1 | 3063 | 1.518 | 0.0251 | No | ||
| 40 | PTK7 | 3303 | 1.432 | 0.0145 | No | ||
| 41 | ERN1 | 3331 | 1.420 | 0.0155 | No | ||
| 42 | TYRO3 | 3347 | 1.413 | 0.0171 | No | ||
| 43 | TTK | 3434 | 1.386 | 0.0147 | No | ||
| 44 | LTK | 3507 | 1.362 | 0.0131 | No | ||
| 45 | RIPK3 | 3560 | 1.346 | 0.0126 | No | ||
| 46 | COASY | 3597 | 1.336 | 0.0129 | No | ||
| 47 | CSNK1D | 3626 | 1.327 | 0.0136 | No | ||
| 48 | MERTK | 3667 | 1.314 | 0.0137 | No | ||
| 49 | RPS6KC1 | 3917 | 1.236 | 0.0022 | No | ||
| 50 | PLK4 | 3925 | 1.235 | 0.0040 | No | ||
| 51 | GTF2H1 | 3951 | 1.227 | 0.0047 | No | ||
| 52 | FGFR4 | 3959 | 1.225 | 0.0064 | No | ||
| 53 | MAP3K6 | 3960 | 1.225 | 0.0085 | No | ||
| 54 | TTN | 3978 | 1.220 | 0.0097 | No | ||
| 55 | CSNK2B | 3994 | 1.216 | 0.0109 | No | ||
| 56 | NRP2 | 4032 | 1.205 | 0.0110 | No | ||
| 57 | STK19 | 4099 | 1.187 | 0.0094 | No | ||
| 58 | CIT | 4246 | 1.147 | 0.0034 | No | ||
| 59 | TESK1 | 4248 | 1.146 | 0.0053 | No | ||
| 60 | MAP2K7 | 4298 | 1.135 | 0.0045 | No | ||
| 61 | BCR | 4306 | 1.134 | 0.0061 | No | ||
| 62 | CSF1R | 4543 | 1.075 | -0.0049 | No | ||
| 63 | EFNA4 | 4620 | 1.056 | -0.0073 | No | ||
| 64 | MARK1 | 4790 | 1.017 | -0.0148 | No | ||
| 65 | KHK | 4840 | 1.004 | -0.0157 | No | ||
| 66 | RPS6KA2 | 4865 | 0.998 | -0.0153 | No | ||
| 67 | MUSK | 4874 | 0.996 | -0.0141 | No | ||
| 68 | CCL2 | 4875 | 0.996 | -0.0124 | No | ||
| 69 | DAPK3 | 4918 | 0.988 | -0.0130 | No | ||
| 70 | MAP3K7 | 5000 | 0.970 | -0.0157 | No | ||
| 71 | STK38 | 5096 | 0.947 | -0.0193 | No | ||
| 72 | MAPK10 | 5114 | 0.942 | -0.0186 | No | ||
| 73 | IRAK3 | 5241 | 0.908 | -0.0239 | No | ||
| 74 | MAPKAPK5 | 5303 | 0.891 | -0.0257 | No | ||
| 75 | PDGFRA | 5317 | 0.887 | -0.0249 | No | ||
| 76 | TSSK2 | 5577 | 0.826 | -0.0377 | No | ||
| 77 | TESK2 | 5709 | 0.797 | -0.0434 | No | ||
| 78 | HUNK | 5726 | 0.792 | -0.0430 | No | ||
| 79 | TEK | 5747 | 0.785 | -0.0427 | No | ||
| 80 | EXOSC10 | 5824 | 0.771 | -0.0455 | No | ||
| 81 | PDGFRL | 5907 | 0.754 | -0.0487 | No | ||
| 82 | BCKDK | 5975 | 0.738 | -0.0511 | No | ||
| 83 | N4BP2 | 5987 | 0.735 | -0.0505 | No | ||
| 84 | PLK1 | 6008 | 0.731 | -0.0503 | No | ||
| 85 | IRAK1 | 6113 | 0.708 | -0.0548 | No | ||
| 86 | MAPK13 | 6168 | 0.698 | -0.0565 | No | ||
| 87 | PDK3 | 6193 | 0.691 | -0.0567 | No | ||
| 88 | FGFR1 | 6252 | 0.677 | -0.0587 | No | ||
| 89 | XYLB | 6259 | 0.675 | -0.0578 | No | ||
| 90 | PNKP | 6267 | 0.674 | -0.0571 | No | ||
| 91 | BRSK2 | 6299 | 0.669 | -0.0576 | No | ||
| 92 | PTK6 | 6329 | 0.661 | -0.0581 | No | ||
| 93 | ROCK1 | 6347 | 0.656 | -0.0579 | No | ||
| 94 | PRKAG1 | 6428 | 0.642 | -0.0611 | No | ||
| 95 | TIE1 | 6522 | 0.624 | -0.0652 | No | ||
| 96 | CSNK2A2 | 6537 | 0.621 | -0.0649 | No | ||
| 97 | RPS6KB2 | 6565 | 0.616 | -0.0653 | No | ||
| 98 | VRK2 | 6598 | 0.609 | -0.0660 | No | ||
| 99 | TWF1 | 6845 | 0.558 | -0.0785 | No | ||
| 100 | AKT1 | 6886 | 0.551 | -0.0797 | No | ||
| 101 | MAP3K9 | 6919 | 0.544 | -0.0805 | No | ||
| 102 | LIMK1 | 6995 | 0.530 | -0.0837 | No | ||
| 103 | PHKG2 | 7072 | 0.517 | -0.0870 | No | ||
| 104 | MAST1 | 7115 | 0.508 | -0.0884 | No | ||
| 105 | EPHA5 | 7146 | 0.502 | -0.0892 | No | ||
| 106 | ROR2 | 7248 | 0.476 | -0.0939 | No | ||
| 107 | NEK2 | 7349 | 0.455 | -0.0985 | No | ||
| 108 | PIK3C2A | 7406 | 0.444 | -0.1008 | No | ||
| 109 | EFNA3 | 7488 | 0.427 | -0.1045 | No | ||
| 110 | CHKA | 7511 | 0.423 | -0.1050 | No | ||
| 111 | CDK7 | 7589 | 0.409 | -0.1085 | No | ||
| 112 | EFNB3 | 7863 | 0.352 | -0.1228 | No | ||
| 113 | TSSK6 | 8108 | 0.301 | -0.1356 | No | ||
| 114 | DAPK2 | 8178 | 0.287 | -0.1389 | No | ||
| 115 | MAPKAPK3 | 8256 | 0.270 | -0.1426 | No | ||
| 116 | ACVR2A | 8372 | 0.250 | -0.1485 | No | ||
| 117 | ACVR1B | 8440 | 0.237 | -0.1517 | No | ||
| 118 | PRKCZ | 8446 | 0.236 | -0.1516 | No | ||
| 119 | TXK | 8749 | 0.174 | -0.1678 | No | ||
| 120 | ACVR1C | 8891 | 0.147 | -0.1752 | No | ||
| 121 | TLK1 | 9043 | 0.117 | -0.1832 | No | ||
| 122 | MAP3K10 | 9073 | 0.112 | -0.1846 | No | ||
| 123 | CHUK | 9136 | 0.102 | -0.1878 | No | ||
| 124 | MAP2K1 | 9431 | 0.042 | -0.2038 | No | ||
| 125 | FRK | 9494 | 0.028 | -0.2071 | No | ||
| 126 | ACVR1 | 9653 | -0.003 | -0.2157 | No | ||
| 127 | NTRK1 | 9683 | -0.009 | -0.2173 | No | ||
| 128 | PIK3CA | 9878 | -0.045 | -0.2278 | No | ||
| 129 | MAPK4 | 10018 | -0.073 | -0.2353 | No | ||
| 130 | WNK3 | 10069 | -0.085 | -0.2379 | No | ||
| 131 | ACVRL1 | 10103 | -0.091 | -0.2395 | No | ||
| 132 | MAP3K13 | 10237 | -0.118 | -0.2466 | No | ||
| 133 | KDR | 10240 | -0.118 | -0.2465 | No | ||
| 134 | TAOK2 | 10248 | -0.120 | -0.2466 | No | ||
| 135 | MAP2K3 | 10511 | -0.169 | -0.2607 | No | ||
| 136 | ERN2 | 10560 | -0.181 | -0.2630 | No | ||
| 137 | HK1 | 10601 | -0.191 | -0.2648 | No | ||
| 138 | SGK2 | 10617 | -0.195 | -0.2653 | No | ||
| 139 | SRPK2 | 10738 | -0.222 | -0.2715 | No | ||
| 140 | TSSK3 | 11026 | -0.283 | -0.2866 | No | ||
| 141 | PDXK | 11284 | -0.335 | -0.3001 | No | ||
| 142 | VRK1 | 11409 | -0.363 | -0.3062 | No | ||
| 143 | MAP3K4 | 11463 | -0.376 | -0.3085 | No | ||
| 144 | GCK | 11905 | -0.472 | -0.3317 | No | ||
| 145 | MKNK2 | 12122 | -0.517 | -0.3426 | No | ||
| 146 | PRKCE | 12144 | -0.521 | -0.3429 | No | ||
| 147 | ERBB4 | 12162 | -0.525 | -0.3429 | No | ||
| 148 | ERBB2 | 12170 | -0.526 | -0.3424 | No | ||
| 149 | PIP5K3 | 12201 | -0.533 | -0.3431 | No | ||
| 150 | MST1R | 12208 | -0.535 | -0.3426 | No | ||
| 151 | EIF2AK4 | 12209 | -0.535 | -0.3416 | No | ||
| 152 | PBK | 12217 | -0.537 | -0.3411 | No | ||
| 153 | PFKP | 12424 | -0.586 | -0.3513 | No | ||
| 154 | PFKFB1 | 12465 | -0.598 | -0.3525 | No | ||
| 155 | PKLR | 12534 | -0.613 | -0.3552 | No | ||
| 156 | CSNK2A1 | 12552 | -0.617 | -0.3550 | No | ||
| 157 | MAPK7 | 12748 | -0.663 | -0.3645 | No | ||
| 158 | STK16 | 12834 | -0.682 | -0.3680 | No | ||
| 159 | FLT3 | 12882 | -0.693 | -0.3694 | No | ||
| 160 | TRIB1 | 12966 | -0.713 | -0.3727 | No | ||
| 161 | KIT | 13043 | -0.731 | -0.3756 | No | ||
| 162 | EGFR | 13064 | -0.736 | -0.3754 | No | ||
| 163 | PRKCI | 13267 | -0.791 | -0.3851 | No | ||
| 164 | CCL4 | 13467 | -0.840 | -0.3945 | No | ||
| 165 | PAK2 | 13592 | -0.871 | -0.3998 | No | ||
| 166 | NEK11 | 13594 | -0.872 | -0.3984 | No | ||
| 167 | CDC42BPA | 13817 | -0.932 | -0.4089 | No | ||
| 168 | TRIM27 | 13862 | -0.945 | -0.4097 | No | ||
| 169 | MINK1 | 13903 | -0.959 | -0.4102 | No | ||
| 170 | MET | 13922 | -0.967 | -0.4096 | No | ||
| 171 | EIF2AK1 | 13989 | -0.984 | -0.4115 | No | ||
| 172 | STK38L | 14219 | -1.051 | -0.4222 | No | ||
| 173 | PRKACB | 14256 | -1.060 | -0.4223 | No | ||
| 174 | ALK | 14320 | -1.081 | -0.4239 | No | ||
| 175 | CDKL1 | 14336 | -1.085 | -0.4229 | No | ||
| 176 | TSSK1A | 14358 | -1.091 | -0.4222 | No | ||
| 177 | MAP3K2 | 14359 | -1.091 | -0.4203 | No | ||
| 178 | MAPK6 | 14362 | -1.092 | -0.4185 | No | ||
| 179 | SPHK1 | 14382 | -1.098 | -0.4177 | No | ||
| 180 | TGFBR1 | 14475 | -1.126 | -0.4208 | No | ||
| 181 | ERBB3 | 14614 | -1.169 | -0.4263 | No | ||
| 182 | AXL | 14846 | -1.255 | -0.4368 | No | ||
| 183 | PAK1 | 14871 | -1.265 | -0.4359 | No | ||
| 184 | ILK | 14888 | -1.271 | -0.4346 | No | ||
| 185 | MARK4 | 15005 | -1.319 | -0.4387 | No | ||
| 186 | MAPK8 | 15028 | -1.330 | -0.4376 | No | ||
| 187 | GALK2 | 15083 | -1.352 | -0.4383 | No | ||
| 188 | PIK3CG | 15103 | -1.358 | -0.4370 | No | ||
| 189 | WNK4 | 15213 | -1.406 | -0.4405 | No | ||
| 190 | IRAK2 | 15218 | -1.408 | -0.4384 | No | ||
| 191 | HIPK3 | 15322 | -1.453 | -0.4415 | No | ||
| 192 | CDK3 | 15389 | -1.489 | -0.4426 | Yes | ||
| 193 | MAPK14 | 15392 | -1.490 | -0.4401 | Yes | ||
| 194 | MAPK12 | 15410 | -1.504 | -0.4385 | Yes | ||
| 195 | PIK3C3 | 15418 | -1.509 | -0.4363 | Yes | ||
| 196 | ATM | 15455 | -1.529 | -0.4356 | Yes | ||
| 197 | MAP4K1 | 15494 | -1.551 | -0.4351 | Yes | ||
| 198 | PHKA1 | 15506 | -1.559 | -0.4330 | Yes | ||
| 199 | CPNE3 | 15533 | -1.574 | -0.4317 | Yes | ||
| 200 | CDK10 | 15545 | -1.583 | -0.4296 | Yes | ||
| 201 | CLK1 | 15599 | -1.608 | -0.4298 | Yes | ||
| 202 | PRKCB1 | 15618 | -1.616 | -0.4280 | Yes | ||
| 203 | STK17B | 15768 | -1.700 | -0.4332 | Yes | ||
| 204 | PIK3CB | 15782 | -1.708 | -0.4310 | Yes | ||
| 205 | UHMK1 | 15785 | -1.710 | -0.4282 | Yes | ||
| 206 | MYLK | 15792 | -1.714 | -0.4256 | Yes | ||
| 207 | PDK1 | 15809 | -1.722 | -0.4235 | Yes | ||
| 208 | DGKZ | 15838 | -1.738 | -0.4221 | Yes | ||
| 209 | RIPK2 | 15887 | -1.764 | -0.4217 | Yes | ||
| 210 | MAP3K5 | 15909 | -1.779 | -0.4198 | Yes | ||
| 211 | PHKA2 | 15944 | -1.805 | -0.4186 | Yes | ||
| 212 | MAPK1 | 15982 | -1.834 | -0.4174 | Yes | ||
| 213 | STK11 | 16037 | -1.869 | -0.4172 | Yes | ||
| 214 | NLK | 16066 | -1.886 | -0.4155 | Yes | ||
| 215 | CSNK1A1 | 16088 | -1.901 | -0.4134 | Yes | ||
| 216 | NEK6 | 16221 | -1.999 | -0.4172 | Yes | ||
| 217 | MAP4K5 | 16226 | -2.001 | -0.4140 | Yes | ||
| 218 | INSR | 16269 | -2.034 | -0.4128 | Yes | ||
| 219 | CAMKK2 | 16332 | -2.086 | -0.4126 | Yes | ||
| 220 | ACVR2B | 16359 | -2.103 | -0.4105 | Yes | ||
| 221 | PRKCH | 16424 | -2.159 | -0.4103 | Yes | ||
| 222 | PRKD1 | 16464 | -2.190 | -0.4086 | Yes | ||
| 223 | MKNK1 | 16480 | -2.203 | -0.4057 | Yes | ||
| 224 | CRIM1 | 16549 | -2.253 | -0.4056 | Yes | ||
| 225 | PXK | 16553 | -2.258 | -0.4019 | Yes | ||
| 226 | AKT2 | 16584 | -2.289 | -0.3996 | Yes | ||
| 227 | PLK3 | 16606 | -2.308 | -0.3968 | Yes | ||
| 228 | LIMK2 | 16635 | -2.331 | -0.3943 | Yes | ||
| 229 | MAP4K4 | 16650 | -2.348 | -0.3911 | Yes | ||
| 230 | EPHB6 | 16688 | -2.383 | -0.3890 | Yes | ||
| 231 | TRIB2 | 16696 | -2.391 | -0.3853 | Yes | ||
| 232 | PINK1 | 16818 | -2.518 | -0.3876 | Yes | ||
| 233 | LMTK2 | 16862 | -2.568 | -0.3856 | Yes | ||
| 234 | STK4 | 16872 | -2.580 | -0.3817 | Yes | ||
| 235 | MAP3K11 | 16964 | -2.684 | -0.3821 | Yes | ||
| 236 | PRKD3 | 16992 | -2.712 | -0.3789 | Yes | ||
| 237 | COL4A3BP | 17051 | -2.787 | -0.3773 | Yes | ||
| 238 | CDK2 | 17094 | -2.849 | -0.3747 | Yes | ||
| 239 | WNK1 | 17096 | -2.850 | -0.3699 | Yes | ||
| 240 | PRKX | 17128 | -2.891 | -0.3667 | Yes | ||
| 241 | MAPK9 | 17129 | -2.896 | -0.3617 | Yes | ||
| 242 | GSK3B | 17139 | -2.906 | -0.3572 | Yes | ||
| 243 | TLK2 | 17145 | -2.916 | -0.3525 | Yes | ||
| 244 | STK10 | 17242 | -3.070 | -0.3525 | Yes | ||
| 245 | DGKE | 17277 | -3.119 | -0.3491 | Yes | ||
| 246 | CSNK1G2 | 17295 | -3.137 | -0.3446 | Yes | ||
| 247 | ULK1 | 17368 | -3.262 | -0.3430 | Yes | ||
| 248 | TNK2 | 17436 | -3.383 | -0.3409 | Yes | ||
| 249 | PIM2 | 17476 | -3.462 | -0.3371 | Yes | ||
| 250 | IGF1R | 17477 | -3.463 | -0.3312 | Yes | ||
| 251 | IGF2R | 17484 | -3.475 | -0.3256 | Yes | ||
| 252 | MAP4K3 | 17566 | -3.626 | -0.3238 | Yes | ||
| 253 | EIF2AK3 | 17575 | -3.644 | -0.3180 | Yes | ||
| 254 | MAP3K8 | 17644 | -3.789 | -0.3152 | Yes | ||
| 255 | CLK2 | 17661 | -3.812 | -0.3096 | Yes | ||
| 256 | ALS2CR2 | 17662 | -3.813 | -0.3031 | Yes | ||
| 257 | PI4K2A | 17668 | -3.832 | -0.2968 | Yes | ||
| 258 | CDK5 | 17776 | -4.065 | -0.2957 | Yes | ||
| 259 | MAP3K12 | 17778 | -4.065 | -0.2888 | Yes | ||
| 260 | SRPK1 | 17868 | -4.293 | -0.2863 | Yes | ||
| 261 | MAPK11 | 17912 | -4.438 | -0.2811 | Yes | ||
| 262 | MAP2K6 | 17919 | -4.460 | -0.2738 | Yes | ||
| 263 | SNF1LK | 17954 | -4.564 | -0.2679 | Yes | ||
| 264 | DGKQ | 17976 | -4.621 | -0.2611 | Yes | ||
| 265 | CRKL | 17994 | -4.686 | -0.2540 | Yes | ||
| 266 | SNF1LK2 | 18018 | -4.752 | -0.2472 | Yes | ||
| 267 | MAP3K3 | 18061 | -4.882 | -0.2411 | Yes | ||
| 268 | PDK2 | 18065 | -4.903 | -0.2329 | Yes | ||
| 269 | PIM1 | 18097 | -5.045 | -0.2260 | Yes | ||
| 270 | BRDT | 18136 | -5.200 | -0.2192 | Yes | ||
| 271 | PFKFB2 | 18163 | -5.386 | -0.2114 | Yes | ||
| 272 | IKBKE | 18185 | -5.485 | -0.2032 | Yes | ||
| 273 | BRD2 | 18187 | -5.490 | -0.1938 | Yes | ||
| 274 | MARK2 | 18200 | -5.546 | -0.1850 | Yes | ||
| 275 | PDPK1 | 18265 | -5.919 | -0.1784 | Yes | ||
| 276 | ITPKB | 18271 | -5.983 | -0.1685 | Yes | ||
| 277 | CSNK1E | 18290 | -6.103 | -0.1590 | Yes | ||
| 278 | DYRK1A | 18303 | -6.167 | -0.1491 | Yes | ||
| 279 | TGFBR2 | 18307 | -6.182 | -0.1387 | Yes | ||
| 280 | SNRK | 18324 | -6.276 | -0.1289 | Yes | ||
| 281 | MAP4K2 | 18412 | -7.157 | -0.1214 | Yes | ||
| 282 | ITK | 18446 | -7.525 | -0.1104 | Yes | ||
| 283 | CDK5R1 | 18479 | -8.062 | -0.0983 | Yes | ||
| 284 | SGK3 | 18490 | -8.275 | -0.0847 | Yes | ||
| 285 | DGKA | 18533 | -9.632 | -0.0706 | Yes | ||
| 286 | PTK2B | 18539 | -9.957 | -0.0538 | Yes | ||
| 287 | TEC | 18587 | -15.689 | -0.0296 | Yes | ||
| 288 | DGKG | 18597 | -18.226 | 0.0010 | Yes |
