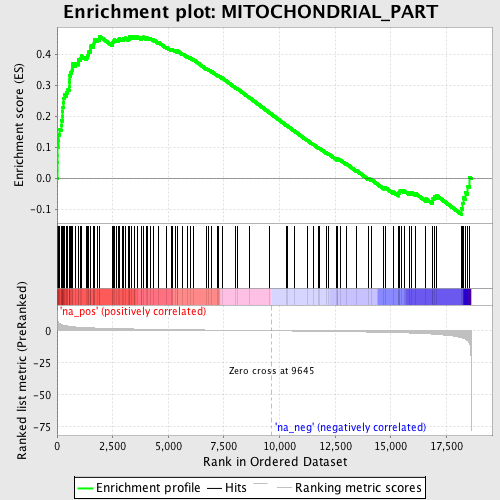
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.45817932 |
| Normalized Enrichment Score (NES) | 1.9722961 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0086406 |
| FWER p-Value | 0.135 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PHB2 | 29 | 7.701 | 0.0247 | Yes | ||
| 2 | TIMM50 | 30 | 7.513 | 0.0503 | Yes | ||
| 3 | ABCB6 | 35 | 7.229 | 0.0747 | Yes | ||
| 4 | BAX | 38 | 6.993 | 0.0984 | Yes | ||
| 5 | TIMM10 | 44 | 6.849 | 0.1214 | Yes | ||
| 6 | MRPS10 | 59 | 6.411 | 0.1425 | Yes | ||
| 7 | TIMM13 | 118 | 5.483 | 0.1581 | Yes | ||
| 8 | IMMT | 182 | 4.852 | 0.1712 | Yes | ||
| 9 | ATP5G1 | 202 | 4.726 | 0.1863 | Yes | ||
| 10 | SLC25A3 | 234 | 4.571 | 0.2001 | Yes | ||
| 11 | NDUFV1 | 241 | 4.535 | 0.2153 | Yes | ||
| 12 | MRPS15 | 259 | 4.452 | 0.2295 | Yes | ||
| 13 | ETFB | 281 | 4.358 | 0.2432 | Yes | ||
| 14 | MRPS28 | 288 | 4.334 | 0.2577 | Yes | ||
| 15 | MRPL55 | 322 | 4.235 | 0.2703 | Yes | ||
| 16 | MRPL12 | 419 | 3.849 | 0.2782 | Yes | ||
| 17 | ATP5G2 | 485 | 3.705 | 0.2873 | Yes | ||
| 18 | ETFA | 540 | 3.558 | 0.2965 | Yes | ||
| 19 | SUPV3L1 | 547 | 3.543 | 0.3083 | Yes | ||
| 20 | BCS1L | 563 | 3.501 | 0.3194 | Yes | ||
| 21 | UQCRC1 | 568 | 3.493 | 0.3311 | Yes | ||
| 22 | ABCF2 | 615 | 3.369 | 0.3401 | Yes | ||
| 23 | MRPS18A | 668 | 3.244 | 0.3483 | Yes | ||
| 24 | AIFM2 | 683 | 3.215 | 0.3585 | Yes | ||
| 25 | MRPS11 | 695 | 3.185 | 0.3687 | Yes | ||
| 26 | TOMM22 | 836 | 2.951 | 0.3712 | Yes | ||
| 27 | COX15 | 949 | 2.798 | 0.3747 | Yes | ||
| 28 | MRPS35 | 964 | 2.787 | 0.3834 | Yes | ||
| 29 | DBT | 1063 | 2.689 | 0.3873 | Yes | ||
| 30 | ATP5E | 1097 | 2.652 | 0.3945 | Yes | ||
| 31 | ABCB7 | 1335 | 2.414 | 0.3899 | Yes | ||
| 32 | SLC25A1 | 1379 | 2.378 | 0.3957 | Yes | ||
| 33 | CYCS | 1401 | 2.360 | 0.4026 | Yes | ||
| 34 | MRPS18C | 1428 | 2.337 | 0.4092 | Yes | ||
| 35 | OPA1 | 1488 | 2.293 | 0.4138 | Yes | ||
| 36 | ATP5G3 | 1512 | 2.282 | 0.4203 | Yes | ||
| 37 | SURF1 | 1521 | 2.278 | 0.4276 | Yes | ||
| 38 | VDAC2 | 1627 | 2.208 | 0.4295 | Yes | ||
| 39 | NDUFS7 | 1656 | 2.186 | 0.4354 | Yes | ||
| 40 | TIMM17A | 1661 | 2.183 | 0.4426 | Yes | ||
| 41 | PMPCA | 1693 | 2.163 | 0.4483 | Yes | ||
| 42 | VDAC3 | 1826 | 2.080 | 0.4483 | Yes | ||
| 43 | MRPS24 | 1892 | 2.043 | 0.4517 | Yes | ||
| 44 | VDAC1 | 1902 | 2.039 | 0.4582 | Yes | ||
| 45 | MRPS12 | 2483 | 1.749 | 0.4328 | No | ||
| 46 | TFB2M | 2506 | 1.737 | 0.4375 | No | ||
| 47 | ATP5D | 2522 | 1.729 | 0.4426 | No | ||
| 48 | TIMM23 | 2566 | 1.710 | 0.4461 | No | ||
| 49 | COX6B2 | 2679 | 1.655 | 0.4457 | No | ||
| 50 | ACN9 | 2756 | 1.628 | 0.4471 | No | ||
| 51 | ATP5F1 | 2805 | 1.609 | 0.4500 | No | ||
| 52 | NDUFAB1 | 2929 | 1.565 | 0.4486 | No | ||
| 53 | ATP5C1 | 2990 | 1.541 | 0.4507 | No | ||
| 54 | UQCRB | 3055 | 1.521 | 0.4524 | No | ||
| 55 | GATM | 3191 | 1.470 | 0.4501 | No | ||
| 56 | NR3C1 | 3247 | 1.453 | 0.4521 | No | ||
| 57 | HSD3B2 | 3258 | 1.449 | 0.4564 | No | ||
| 58 | SDHD | 3348 | 1.413 | 0.4564 | No | ||
| 59 | MRPL10 | 3481 | 1.370 | 0.4540 | No | ||
| 60 | MRPL51 | 3496 | 1.366 | 0.4579 | No | ||
| 61 | NDUFA6 | 3615 | 1.331 | 0.4560 | No | ||
| 62 | NFS1 | 3789 | 1.281 | 0.4510 | No | ||
| 63 | NDUFS2 | 3794 | 1.278 | 0.4552 | No | ||
| 64 | RAB11FIP5 | 3868 | 1.252 | 0.4555 | No | ||
| 65 | UCP3 | 3995 | 1.216 | 0.4528 | No | ||
| 66 | ATP5O | 4080 | 1.191 | 0.4523 | No | ||
| 67 | NDUFS8 | 4177 | 1.165 | 0.4511 | No | ||
| 68 | ATP5J | 4340 | 1.126 | 0.4462 | No | ||
| 69 | PIN4 | 4539 | 1.076 | 0.4391 | No | ||
| 70 | MFN2 | 4917 | 0.988 | 0.4221 | No | ||
| 71 | NDUFA1 | 5129 | 0.937 | 0.4139 | No | ||
| 72 | HADHB | 5176 | 0.925 | 0.4145 | No | ||
| 73 | GRPEL1 | 5304 | 0.891 | 0.4107 | No | ||
| 74 | POLG2 | 5412 | 0.862 | 0.4079 | No | ||
| 75 | MAOB | 5417 | 0.860 | 0.4106 | No | ||
| 76 | CS | 5656 | 0.810 | 0.4005 | No | ||
| 77 | ATP5A1 | 5869 | 0.760 | 0.3916 | No | ||
| 78 | BCKDK | 5975 | 0.738 | 0.3884 | No | ||
| 79 | TIMM9 | 6130 | 0.705 | 0.3825 | No | ||
| 80 | MTX2 | 6719 | 0.587 | 0.3527 | No | ||
| 81 | CENTA2 | 6808 | 0.568 | 0.3498 | No | ||
| 82 | HTRA2 | 6949 | 0.541 | 0.3441 | No | ||
| 83 | MRPS16 | 7217 | 0.483 | 0.3313 | No | ||
| 84 | NDUFA9 | 7272 | 0.470 | 0.3300 | No | ||
| 85 | MRPS22 | 7418 | 0.441 | 0.3237 | No | ||
| 86 | MRPL52 | 8032 | 0.319 | 0.2916 | No | ||
| 87 | HCCS | 8128 | 0.297 | 0.2875 | No | ||
| 88 | SLC25A15 | 8638 | 0.197 | 0.2606 | No | ||
| 89 | HSD3B1 | 9529 | 0.020 | 0.2125 | No | ||
| 90 | PITRM1 | 10303 | -0.130 | 0.1712 | No | ||
| 91 | SDHA | 10362 | -0.140 | 0.1685 | No | ||
| 92 | BCKDHB | 10656 | -0.205 | 0.1534 | No | ||
| 93 | BCKDHA | 11247 | -0.328 | 0.1226 | No | ||
| 94 | RHOT1 | 11518 | -0.388 | 0.1093 | No | ||
| 95 | MRPL40 | 11741 | -0.439 | 0.0988 | No | ||
| 96 | CASQ1 | 11793 | -0.450 | 0.0975 | No | ||
| 97 | PPOX | 12093 | -0.511 | 0.0831 | No | ||
| 98 | PHB | 12202 | -0.533 | 0.0791 | No | ||
| 99 | ALDH4A1 | 12570 | -0.621 | 0.0614 | No | ||
| 100 | MRPL23 | 12605 | -0.630 | 0.0617 | No | ||
| 101 | MRPS36 | 12613 | -0.631 | 0.0634 | No | ||
| 102 | SLC25A22 | 12726 | -0.658 | 0.0596 | No | ||
| 103 | BNIP3 | 12999 | -0.719 | 0.0474 | No | ||
| 104 | TIMM17B | 13468 | -0.840 | 0.0249 | No | ||
| 105 | TIMM44 | 13979 | -0.982 | 0.0007 | No | ||
| 106 | TIMM8B | 14121 | -1.019 | -0.0035 | No | ||
| 107 | ATP5B | 14686 | -1.194 | -0.0299 | No | ||
| 108 | DNAJA3 | 14760 | -1.218 | -0.0297 | No | ||
| 109 | NDUFS3 | 15097 | -1.356 | -0.0433 | No | ||
| 110 | MPV17 | 15342 | -1.466 | -0.0515 | No | ||
| 111 | NDUFA2 | 15344 | -1.467 | -0.0466 | No | ||
| 112 | OGDH | 15383 | -1.486 | -0.0435 | No | ||
| 113 | UQCRH | 15411 | -1.506 | -0.0399 | No | ||
| 114 | TOMM34 | 15458 | -1.530 | -0.0372 | No | ||
| 115 | NNT | 15616 | -1.616 | -0.0401 | No | ||
| 116 | FIS1 | 15831 | -1.735 | -0.0458 | No | ||
| 117 | RAF1 | 15947 | -1.805 | -0.0459 | No | ||
| 118 | NDUFS1 | 16100 | -1.909 | -0.0476 | No | ||
| 119 | MRPL32 | 16578 | -2.283 | -0.0656 | No | ||
| 120 | NDUFA13 | 16881 | -2.593 | -0.0731 | No | ||
| 121 | NDUFS4 | 16892 | -2.602 | -0.0648 | No | ||
| 122 | ACADM | 16942 | -2.663 | -0.0584 | No | ||
| 123 | MRPS21 | 17055 | -2.793 | -0.0549 | No | ||
| 124 | RHOT2 | 18183 | -5.480 | -0.0972 | No | ||
| 125 | MCL1 | 18226 | -5.653 | -0.0802 | No | ||
| 126 | ALAS2 | 18249 | -5.792 | -0.0617 | No | ||
| 127 | SLC25A11 | 18345 | -6.504 | -0.0447 | No | ||
| 128 | CASP7 | 18463 | -7.844 | -0.0243 | No | ||
| 129 | BCL2 | 18530 | -9.542 | 0.0047 | No |
