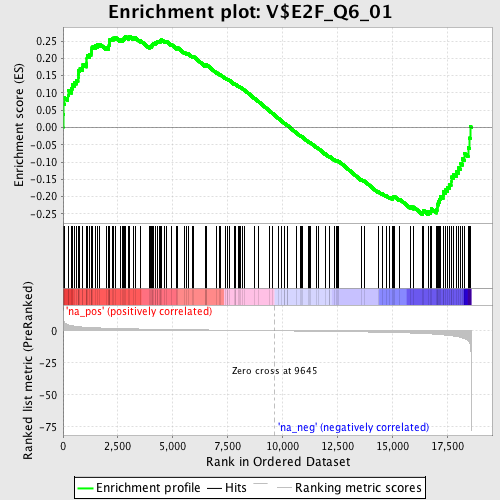
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E2F_Q6_01 |
| Enrichment Score (ES) | 0.26440072 |
| Normalized Enrichment Score (NES) | 1.1714747 |
| Nominal p-value | 0.08866995 |
| FDR q-value | 0.5358943 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BMP7 | 3 | 11.642 | 0.0387 | Yes | ||
| 2 | EIF5A | 17 | 8.994 | 0.0681 | Yes | ||
| 3 | HNRPA1 | 66 | 6.164 | 0.0861 | Yes | ||
| 4 | DNAJC11 | 222 | 4.614 | 0.0931 | Yes | ||
| 5 | PCSK4 | 239 | 4.551 | 0.1075 | Yes | ||
| 6 | RPS19 | 387 | 3.975 | 0.1128 | Yes | ||
| 7 | NUTF2 | 418 | 3.850 | 0.1240 | Yes | ||
| 8 | TMEM4 | 531 | 3.572 | 0.1299 | Yes | ||
| 9 | KLHDC3 | 609 | 3.385 | 0.1370 | Yes | ||
| 10 | PRIM1 | 679 | 3.222 | 0.1441 | Yes | ||
| 11 | CYR61 | 685 | 3.206 | 0.1545 | Yes | ||
| 12 | FANCL | 700 | 3.172 | 0.1644 | Yes | ||
| 13 | ERG | 768 | 3.062 | 0.1710 | Yes | ||
| 14 | UNG | 881 | 2.882 | 0.1745 | Yes | ||
| 15 | NCL | 903 | 2.857 | 0.1829 | Yes | ||
| 16 | ACIN1 | 1051 | 2.702 | 0.1840 | Yes | ||
| 17 | RFC1 | 1084 | 2.666 | 0.1912 | Yes | ||
| 18 | MELK | 1085 | 2.665 | 0.2001 | Yes | ||
| 19 | CDKN1A | 1088 | 2.662 | 0.2089 | Yes | ||
| 20 | CACNA1G | 1202 | 2.554 | 0.2113 | Yes | ||
| 21 | STAG1 | 1273 | 2.475 | 0.2158 | Yes | ||
| 22 | MYC | 1285 | 2.465 | 0.2234 | Yes | ||
| 23 | PVRL1 | 1313 | 2.437 | 0.2301 | Yes | ||
| 24 | ERBB2IP | 1358 | 2.397 | 0.2357 | Yes | ||
| 25 | ATAD2 | 1479 | 2.298 | 0.2369 | Yes | ||
| 26 | RAD51 | 1567 | 2.241 | 0.2397 | Yes | ||
| 27 | MCM4 | 1678 | 2.171 | 0.2410 | Yes | ||
| 28 | SFRS2 | 1972 | 2.002 | 0.2318 | Yes | ||
| 29 | YWHAQ | 2088 | 1.942 | 0.2321 | Yes | ||
| 30 | HMGN2 | 2089 | 1.941 | 0.2385 | Yes | ||
| 31 | LUC7L2 | 2108 | 1.932 | 0.2440 | Yes | ||
| 32 | TLE3 | 2120 | 1.928 | 0.2499 | Yes | ||
| 33 | MCM6 | 2131 | 1.924 | 0.2558 | Yes | ||
| 34 | TNPO2 | 2229 | 1.870 | 0.2568 | Yes | ||
| 35 | LIG1 | 2301 | 1.838 | 0.2591 | Yes | ||
| 36 | PHF15 | 2394 | 1.798 | 0.2601 | Yes | ||
| 37 | SLITRK3 | 2621 | 1.685 | 0.2535 | Yes | ||
| 38 | PRKCSH | 2718 | 1.640 | 0.2538 | Yes | ||
| 39 | CLPTM1 | 2771 | 1.622 | 0.2564 | Yes | ||
| 40 | HOXA9 | 2809 | 1.606 | 0.2597 | Yes | ||
| 41 | AP4M1 | 2838 | 1.597 | 0.2635 | Yes | ||
| 42 | HTF9C | 2998 | 1.539 | 0.2601 | Yes | ||
| 43 | RPS6KA5 | 3014 | 1.536 | 0.2644 | Yes | ||
| 44 | STAG2 | 3198 | 1.469 | 0.2594 | No | ||
| 45 | APRIN | 3276 | 1.441 | 0.2600 | No | ||
| 46 | GRIA4 | 3518 | 1.357 | 0.2515 | No | ||
| 47 | PLK4 | 3925 | 1.235 | 0.2337 | No | ||
| 48 | MCM7 | 3963 | 1.224 | 0.2358 | No | ||
| 49 | NRP2 | 4032 | 1.205 | 0.2361 | No | ||
| 50 | MCM3 | 4053 | 1.199 | 0.2390 | No | ||
| 51 | CDC45L | 4089 | 1.188 | 0.2411 | No | ||
| 52 | XPO5 | 4104 | 1.186 | 0.2443 | No | ||
| 53 | RBBP4 | 4202 | 1.157 | 0.2429 | No | ||
| 54 | SUV39H1 | 4210 | 1.155 | 0.2464 | No | ||
| 55 | RANBP1 | 4279 | 1.139 | 0.2465 | No | ||
| 56 | GMNN | 4284 | 1.138 | 0.2501 | No | ||
| 57 | KCND2 | 4374 | 1.118 | 0.2490 | No | ||
| 58 | ADAMTS2 | 4422 | 1.106 | 0.2502 | No | ||
| 59 | NASP | 4443 | 1.100 | 0.2528 | No | ||
| 60 | FBXO5 | 4464 | 1.095 | 0.2553 | No | ||
| 61 | E2F1 | 4642 | 1.052 | 0.2493 | No | ||
| 62 | SEMA5A | 4715 | 1.034 | 0.2488 | No | ||
| 63 | POLD1 | 4943 | 0.982 | 0.2398 | No | ||
| 64 | SFMBT1 | 5187 | 0.920 | 0.2297 | No | ||
| 65 | EZH2 | 5215 | 0.913 | 0.2313 | No | ||
| 66 | BAI1 | 5518 | 0.839 | 0.2178 | No | ||
| 67 | E2F3 | 5630 | 0.814 | 0.2145 | No | ||
| 68 | POLE2 | 5700 | 0.799 | 0.2134 | No | ||
| 69 | SALL1 | 5900 | 0.755 | 0.2051 | No | ||
| 70 | REPS2 | 5945 | 0.746 | 0.2052 | No | ||
| 71 | ARID4A | 6489 | 0.631 | 0.1779 | No | ||
| 72 | DLST | 6498 | 0.629 | 0.1796 | No | ||
| 73 | KPNB1 | 6534 | 0.622 | 0.1798 | No | ||
| 74 | CDC6 | 6535 | 0.622 | 0.1819 | No | ||
| 75 | HNRPD | 6999 | 0.530 | 0.1586 | No | ||
| 76 | ACBD6 | 7126 | 0.506 | 0.1534 | No | ||
| 77 | STMN1 | 7185 | 0.491 | 0.1519 | No | ||
| 78 | DLL4 | 7398 | 0.445 | 0.1419 | No | ||
| 79 | PAQR4 | 7474 | 0.430 | 0.1393 | No | ||
| 80 | NDUFA11 | 7499 | 0.425 | 0.1394 | No | ||
| 81 | DNMT1 | 7574 | 0.412 | 0.1368 | No | ||
| 82 | MCM2 | 7792 | 0.369 | 0.1263 | No | ||
| 83 | DUSP7 | 7838 | 0.358 | 0.1250 | No | ||
| 84 | DCLRE1A | 7874 | 0.350 | 0.1243 | No | ||
| 85 | TOPBP1 | 7998 | 0.326 | 0.1187 | No | ||
| 86 | HIST1H1D | 8038 | 0.318 | 0.1177 | No | ||
| 87 | CDC25A | 8094 | 0.305 | 0.1157 | No | ||
| 88 | SNCB | 8154 | 0.293 | 0.1135 | No | ||
| 89 | PODN | 8277 | 0.265 | 0.1078 | No | ||
| 90 | NKX2-2 | 8721 | 0.180 | 0.0844 | No | ||
| 91 | CDCA7 | 8907 | 0.143 | 0.0749 | No | ||
| 92 | BRMS1L | 9416 | 0.046 | 0.0475 | No | ||
| 93 | UFD1L | 9544 | 0.017 | 0.0407 | No | ||
| 94 | RBL1 | 9838 | -0.036 | 0.0249 | No | ||
| 95 | TBX3 | 9958 | -0.062 | 0.0187 | No | ||
| 96 | BARHL1 | 10097 | -0.090 | 0.0115 | No | ||
| 97 | SLC16A2 | 10111 | -0.093 | 0.0111 | No | ||
| 98 | MAP3K13 | 10237 | -0.118 | 0.0048 | No | ||
| 99 | GPRC5B | 10245 | -0.120 | 0.0048 | No | ||
| 100 | PTMA | 10659 | -0.206 | -0.0169 | No | ||
| 101 | H2AFZ | 10801 | -0.236 | -0.0238 | No | ||
| 102 | EPHB1 | 10857 | -0.246 | -0.0259 | No | ||
| 103 | IPO7 | 10891 | -0.255 | -0.0268 | No | ||
| 104 | SFRS1 | 11161 | -0.311 | -0.0404 | No | ||
| 105 | HTR2C | 11216 | -0.322 | -0.0422 | No | ||
| 106 | GLRA3 | 11269 | -0.333 | -0.0439 | No | ||
| 107 | ADAMTS9 | 11548 | -0.393 | -0.0577 | No | ||
| 108 | PRKDC | 11625 | -0.410 | -0.0604 | No | ||
| 109 | WEE1 | 11945 | -0.478 | -0.0761 | No | ||
| 110 | ETV4 | 12130 | -0.519 | -0.0843 | No | ||
| 111 | CASP8AP2 | 12131 | -0.519 | -0.0826 | No | ||
| 112 | FMO4 | 12366 | -0.570 | -0.0934 | No | ||
| 113 | CTCF | 12473 | -0.599 | -0.0971 | No | ||
| 114 | ORC1L | 12488 | -0.604 | -0.0959 | No | ||
| 115 | DMRT1 | 12566 | -0.621 | -0.0979 | No | ||
| 116 | PEG3 | 13585 | -0.869 | -0.1502 | No | ||
| 117 | MAT2A | 13726 | -0.905 | -0.1547 | No | ||
| 118 | KCNA6 | 14371 | -1.096 | -0.1860 | No | ||
| 119 | FLI1 | 14542 | -1.147 | -0.1913 | No | ||
| 120 | DMD | 14721 | -1.204 | -0.1969 | No | ||
| 121 | FANCG | 14896 | -1.273 | -0.2021 | No | ||
| 122 | SLC6A4 | 15001 | -1.317 | -0.2033 | No | ||
| 123 | ANP32E | 15036 | -1.333 | -0.2007 | No | ||
| 124 | PCNA | 15099 | -1.356 | -0.1996 | No | ||
| 125 | PCDH7 | 15341 | -1.466 | -0.2077 | No | ||
| 126 | BCOR | 15828 | -1.731 | -0.2283 | No | ||
| 127 | SLITRK4 | 15975 | -1.828 | -0.2301 | No | ||
| 128 | RRM2 | 16378 | -2.120 | -0.2447 | No | ||
| 129 | MXD3 | 16408 | -2.146 | -0.2391 | No | ||
| 130 | RALY | 16640 | -2.336 | -0.2438 | No | ||
| 131 | CITED2 | 16757 | -2.461 | -0.2419 | No | ||
| 132 | MAZ | 16783 | -2.485 | -0.2350 | No | ||
| 133 | UCHL1 | 17018 | -2.743 | -0.2385 | No | ||
| 134 | DAXX | 17045 | -2.781 | -0.2306 | No | ||
| 135 | CBX5 | 17049 | -2.784 | -0.2214 | No | ||
| 136 | PPM1D | 17106 | -2.862 | -0.2149 | No | ||
| 137 | NHLRC2 | 17167 | -2.941 | -0.2083 | No | ||
| 138 | TIAM1 | 17194 | -2.990 | -0.1997 | No | ||
| 139 | ZCCHC8 | 17318 | -3.172 | -0.1958 | No | ||
| 140 | POLH | 17336 | -3.211 | -0.1860 | No | ||
| 141 | ATE1 | 17443 | -3.402 | -0.1804 | No | ||
| 142 | CASP2 | 17527 | -3.563 | -0.1729 | No | ||
| 143 | FBXL20 | 17616 | -3.714 | -0.1653 | No | ||
| 144 | DDB2 | 17682 | -3.864 | -0.1559 | No | ||
| 145 | KLF5 | 17688 | -3.882 | -0.1432 | No | ||
| 146 | AXUD1 | 17786 | -4.073 | -0.1348 | No | ||
| 147 | USP52 | 17922 | -4.469 | -0.1272 | No | ||
| 148 | DPYSL2 | 18025 | -4.769 | -0.1168 | No | ||
| 149 | PPP1R9B | 18121 | -5.135 | -0.1048 | No | ||
| 150 | E2F7 | 18197 | -5.528 | -0.0904 | No | ||
| 151 | RAB11B | 18285 | -6.081 | -0.0748 | No | ||
| 152 | DCK | 18459 | -7.779 | -0.0581 | No | ||
| 153 | HRBL | 18523 | -9.325 | -0.0304 | No | ||
| 154 | DUSP6 | 18554 | -10.586 | 0.0034 | No |
