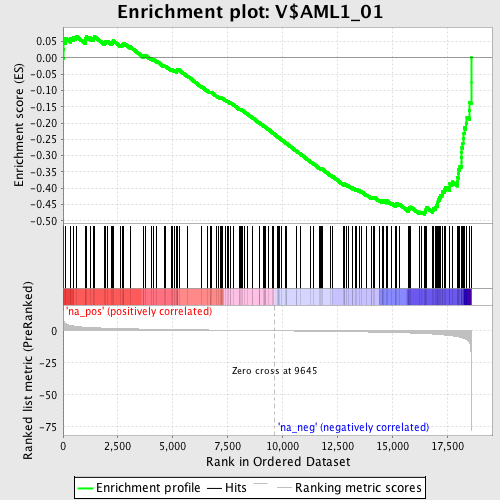
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$AML1_01 |
| Enrichment Score (ES) | -0.48044345 |
| Normalized Enrichment Score (NES) | -1.7864279 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08595707 |
| FWER p-Value | 0.61 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | INPPL1 | 8 | 9.461 | 0.0266 | No | ||
| 2 | MTX1 | 24 | 8.334 | 0.0496 | No | ||
| 3 | EIF2B3 | 116 | 5.490 | 0.0603 | No | ||
| 4 | JDP2 | 342 | 4.146 | 0.0599 | No | ||
| 5 | SLC6A9 | 484 | 3.705 | 0.0629 | No | ||
| 6 | SUPT16H | 587 | 3.436 | 0.0672 | No | ||
| 7 | TNFSF4 | 1019 | 2.729 | 0.0516 | No | ||
| 8 | LY9 | 1027 | 2.723 | 0.0590 | No | ||
| 9 | ACIN1 | 1051 | 2.702 | 0.0654 | No | ||
| 10 | ARHGAP21 | 1251 | 2.505 | 0.0618 | No | ||
| 11 | KCTD4 | 1390 | 2.370 | 0.0611 | No | ||
| 12 | WNT8B | 1407 | 2.354 | 0.0670 | No | ||
| 13 | BMF | 1901 | 2.039 | 0.0461 | No | ||
| 14 | HOXC6 | 1944 | 2.013 | 0.0495 | No | ||
| 15 | RAB39 | 2002 | 1.985 | 0.0521 | No | ||
| 16 | BCAR3 | 2193 | 1.886 | 0.0472 | No | ||
| 17 | MAMDC1 | 2254 | 1.859 | 0.0492 | No | ||
| 18 | MPL | 2282 | 1.845 | 0.0530 | No | ||
| 19 | HDGF | 2617 | 1.689 | 0.0398 | No | ||
| 20 | CORO1C | 2685 | 1.652 | 0.0409 | No | ||
| 21 | ATP2A2 | 2760 | 1.626 | 0.0415 | No | ||
| 22 | PRICKLE1 | 2769 | 1.622 | 0.0457 | No | ||
| 23 | EIF2C1 | 3056 | 1.521 | 0.0345 | No | ||
| 24 | SCN8A | 3645 | 1.317 | 0.0064 | No | ||
| 25 | MRPS31 | 3683 | 1.309 | 0.0081 | No | ||
| 26 | THBS3 | 3761 | 1.288 | 0.0076 | No | ||
| 27 | TCF12 | 4014 | 1.210 | -0.0026 | No | ||
| 28 | TNNT2 | 4115 | 1.183 | -0.0046 | No | ||
| 29 | PTPN22 | 4269 | 1.141 | -0.0096 | No | ||
| 30 | COL4A2 | 4599 | 1.062 | -0.0244 | No | ||
| 31 | PSMA1 | 4659 | 1.048 | -0.0246 | No | ||
| 32 | FGF4 | 4931 | 0.984 | -0.0365 | No | ||
| 33 | DAB1 | 4981 | 0.975 | -0.0364 | No | ||
| 34 | GPD1 | 5071 | 0.954 | -0.0385 | No | ||
| 35 | SLC2A3 | 5173 | 0.926 | -0.0413 | No | ||
| 36 | SLC37A2 | 5191 | 0.919 | -0.0396 | No | ||
| 37 | AGTRL1 | 5193 | 0.919 | -0.0371 | No | ||
| 38 | GRK5 | 5209 | 0.915 | -0.0353 | No | ||
| 39 | CRTAC1 | 5282 | 0.897 | -0.0366 | No | ||
| 40 | GTF2A1 | 5684 | 0.803 | -0.0560 | No | ||
| 41 | ZBTB8 | 6306 | 0.668 | -0.0878 | No | ||
| 42 | ADAMTS8 | 6558 | 0.617 | -0.0996 | No | ||
| 43 | DKK1 | 6695 | 0.591 | -0.1053 | No | ||
| 44 | PDZK1 | 6713 | 0.588 | -0.1046 | No | ||
| 45 | ANXA8 | 6754 | 0.581 | -0.1051 | No | ||
| 46 | CRY1 | 6997 | 0.530 | -0.1167 | No | ||
| 47 | PXN | 7089 | 0.511 | -0.1202 | No | ||
| 48 | AKAP3 | 7153 | 0.501 | -0.1221 | No | ||
| 49 | SOX5 | 7196 | 0.489 | -0.1230 | No | ||
| 50 | HOXC4 | 7214 | 0.484 | -0.1226 | No | ||
| 51 | NDUFA9 | 7272 | 0.470 | -0.1243 | No | ||
| 52 | TITF1 | 7410 | 0.444 | -0.1305 | No | ||
| 53 | HIVEP3 | 7476 | 0.430 | -0.1328 | No | ||
| 54 | ADAMTS4 | 7533 | 0.417 | -0.1346 | No | ||
| 55 | ARHGAP12 | 7615 | 0.404 | -0.1379 | No | ||
| 56 | PICALM | 7644 | 0.398 | -0.1382 | No | ||
| 57 | CHES1 | 7768 | 0.375 | -0.1438 | No | ||
| 58 | TLL2 | 8035 | 0.319 | -0.1573 | No | ||
| 59 | PCF11 | 8060 | 0.313 | -0.1577 | No | ||
| 60 | IL23A | 8084 | 0.307 | -0.1581 | No | ||
| 61 | ITGB7 | 8124 | 0.298 | -0.1594 | No | ||
| 62 | TPM1 | 8173 | 0.290 | -0.1612 | No | ||
| 63 | FCER1G | 8259 | 0.270 | -0.1650 | No | ||
| 64 | ANKRD1 | 8421 | 0.243 | -0.1730 | No | ||
| 65 | PDE3B | 8628 | 0.199 | -0.1836 | No | ||
| 66 | ANK3 | 8646 | 0.196 | -0.1840 | No | ||
| 67 | SLC15A3 | 8930 | 0.138 | -0.1989 | No | ||
| 68 | STRC | 8954 | 0.135 | -0.1998 | No | ||
| 69 | COL4A1 | 8964 | 0.132 | -0.1999 | No | ||
| 70 | DNM3 | 9121 | 0.104 | -0.2081 | No | ||
| 71 | SCEL | 9164 | 0.095 | -0.2101 | No | ||
| 72 | CTSK | 9172 | 0.095 | -0.2102 | No | ||
| 73 | KCNJ1 | 9218 | 0.085 | -0.2124 | No | ||
| 74 | PLXNC1 | 9237 | 0.081 | -0.2131 | No | ||
| 75 | NRAS | 9380 | 0.052 | -0.2207 | No | ||
| 76 | MATN1 | 9528 | 0.020 | -0.2286 | No | ||
| 77 | SIRT1 | 9607 | 0.007 | -0.2328 | No | ||
| 78 | RIN1 | 9750 | -0.019 | -0.2404 | No | ||
| 79 | SLITRK5 | 9796 | -0.027 | -0.2428 | No | ||
| 80 | HLX1 | 9815 | -0.030 | -0.2437 | No | ||
| 81 | ABCD4 | 9882 | -0.045 | -0.2471 | No | ||
| 82 | PGF | 9961 | -0.062 | -0.2512 | No | ||
| 83 | TBK1 | 10121 | -0.096 | -0.2595 | No | ||
| 84 | ARHGEF2 | 10171 | -0.104 | -0.2619 | No | ||
| 85 | MMP13 | 10193 | -0.110 | -0.2627 | No | ||
| 86 | NOTCH2 | 10622 | -0.197 | -0.2853 | No | ||
| 87 | ENTPD1 | 10652 | -0.204 | -0.2863 | No | ||
| 88 | COL9A2 | 10813 | -0.238 | -0.2943 | No | ||
| 89 | GRASP | 11295 | -0.339 | -0.3194 | No | ||
| 90 | VRK1 | 11409 | -0.363 | -0.3245 | No | ||
| 91 | RGS18 | 11696 | -0.426 | -0.3388 | No | ||
| 92 | CLDN10 | 11738 | -0.438 | -0.3398 | No | ||
| 93 | LMO2 | 11761 | -0.443 | -0.3397 | No | ||
| 94 | BTG4 | 11800 | -0.452 | -0.3405 | No | ||
| 95 | MKRN3 | 12191 | -0.531 | -0.3601 | No | ||
| 96 | PDE6H | 12298 | -0.555 | -0.3643 | No | ||
| 97 | NR4A1 | 12782 | -0.668 | -0.3885 | No | ||
| 98 | NGB | 12806 | -0.676 | -0.3878 | No | ||
| 99 | ATP6V0B | 12821 | -0.679 | -0.3867 | No | ||
| 100 | NRXN3 | 12908 | -0.699 | -0.3893 | No | ||
| 101 | NUCB2 | 12991 | -0.716 | -0.3917 | No | ||
| 102 | GZMB | 13189 | -0.772 | -0.4002 | No | ||
| 103 | NFRKB | 13195 | -0.773 | -0.3983 | No | ||
| 104 | PAX6 | 13346 | -0.810 | -0.4041 | No | ||
| 105 | TBX5 | 13391 | -0.821 | -0.4041 | No | ||
| 106 | UBL3 | 13498 | -0.848 | -0.4074 | No | ||
| 107 | MYOD1 | 13586 | -0.870 | -0.4097 | No | ||
| 108 | VIM | 13841 | -0.940 | -0.4208 | No | ||
| 109 | EIF3S1 | 14058 | -1.000 | -0.4296 | No | ||
| 110 | SLC37A4 | 14065 | -1.002 | -0.4271 | No | ||
| 111 | RGS1 | 14130 | -1.022 | -0.4276 | No | ||
| 112 | CYP17A1 | 14179 | -1.040 | -0.4273 | No | ||
| 113 | MOAP1 | 14421 | -1.110 | -0.4372 | No | ||
| 114 | FLI1 | 14542 | -1.147 | -0.4404 | No | ||
| 115 | DACT1 | 14551 | -1.149 | -0.4375 | No | ||
| 116 | BLOC1S2 | 14592 | -1.163 | -0.4364 | No | ||
| 117 | LUZP1 | 14757 | -1.216 | -0.4418 | No | ||
| 118 | FGD4 | 14764 | -1.222 | -0.4386 | No | ||
| 119 | SCN3B | 14983 | -1.308 | -0.4467 | No | ||
| 120 | IFNG | 15149 | -1.377 | -0.4517 | No | ||
| 121 | SCNN1A | 15168 | -1.388 | -0.4488 | No | ||
| 122 | FOXD2 | 15189 | -1.397 | -0.4458 | No | ||
| 123 | MR1 | 15346 | -1.468 | -0.4501 | No | ||
| 124 | ITGA10 | 15732 | -1.681 | -0.4662 | No | ||
| 125 | SNX1 | 15761 | -1.698 | -0.4629 | No | ||
| 126 | BATF | 15801 | -1.718 | -0.4601 | No | ||
| 127 | POU2F1 | 15845 | -1.743 | -0.4574 | No | ||
| 128 | RDH5 | 16234 | -2.005 | -0.4727 | No | ||
| 129 | AP1G2 | 16349 | -2.098 | -0.4729 | No | ||
| 130 | RICS | 16489 | -2.213 | -0.4741 | Yes | ||
| 131 | JMJD1C | 16516 | -2.225 | -0.4692 | Yes | ||
| 132 | EDARADD | 16536 | -2.242 | -0.4638 | Yes | ||
| 133 | RHOG | 16566 | -2.270 | -0.4589 | Yes | ||
| 134 | LCK | 16851 | -2.559 | -0.4670 | Yes | ||
| 135 | TACSTD2 | 16884 | -2.594 | -0.4613 | Yes | ||
| 136 | MAP3K11 | 16964 | -2.684 | -0.4579 | Yes | ||
| 137 | ARF3 | 16996 | -2.715 | -0.4519 | Yes | ||
| 138 | DNAJC14 | 17062 | -2.805 | -0.4474 | Yes | ||
| 139 | TACC2 | 17085 | -2.838 | -0.4405 | Yes | ||
| 140 | KIF1B | 17100 | -2.851 | -0.4331 | Yes | ||
| 141 | NHLRC2 | 17167 | -2.941 | -0.4283 | Yes | ||
| 142 | BLOC1S1 | 17206 | -3.009 | -0.4217 | Yes | ||
| 143 | ARNT | 17271 | -3.107 | -0.4163 | Yes | ||
| 144 | RORC | 17304 | -3.150 | -0.4091 | Yes | ||
| 145 | ATF7IP | 17385 | -3.293 | -0.4040 | Yes | ||
| 146 | MMP14 | 17421 | -3.354 | -0.3963 | Yes | ||
| 147 | PTPN7 | 17618 | -3.720 | -0.3963 | Yes | ||
| 148 | DIABLO | 17625 | -3.729 | -0.3860 | Yes | ||
| 149 | CD6 | 17729 | -3.956 | -0.3803 | Yes | ||
| 150 | CREM | 17966 | -4.594 | -0.3800 | Yes | ||
| 151 | S100A9 | 17987 | -4.664 | -0.3678 | Yes | ||
| 152 | CAP1 | 18004 | -4.714 | -0.3552 | Yes | ||
| 153 | EMP1 | 18014 | -4.744 | -0.3421 | Yes | ||
| 154 | HIPK1 | 18085 | -4.982 | -0.3317 | Yes | ||
| 155 | LRP5 | 18149 | -5.303 | -0.3200 | Yes | ||
| 156 | CBL | 18150 | -5.305 | -0.3048 | Yes | ||
| 157 | STAT2 | 18166 | -5.430 | -0.2901 | Yes | ||
| 158 | RAB5B | 18176 | -5.449 | -0.2751 | Yes | ||
| 159 | CUGBP1 | 18217 | -5.616 | -0.2612 | Yes | ||
| 160 | SEMA4A | 18231 | -5.683 | -0.2457 | Yes | ||
| 161 | RAG1 | 18266 | -5.929 | -0.2306 | Yes | ||
| 162 | PACS1 | 18280 | -6.019 | -0.2141 | Yes | ||
| 163 | MADD | 18362 | -6.643 | -0.1995 | Yes | ||
| 164 | CD69 | 18406 | -7.048 | -0.1817 | Yes | ||
| 165 | SLC12A6 | 18516 | -9.060 | -0.1618 | Yes | ||
| 166 | DGKA | 18533 | -9.632 | -0.1351 | Yes | ||
| 167 | EGR2 | 18610 | -22.681 | -0.0745 | Yes | ||
| 168 | XCL1 | 18612 | -26.194 | 0.0002 | Yes |