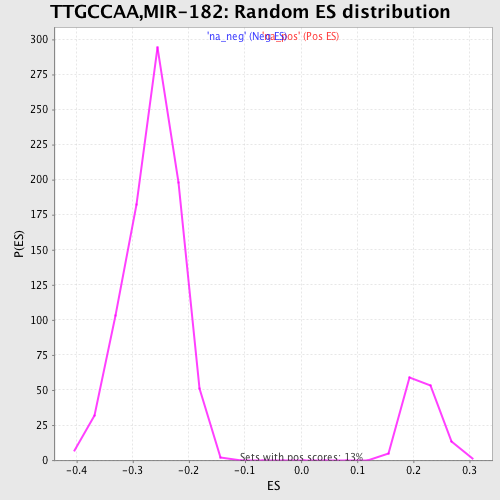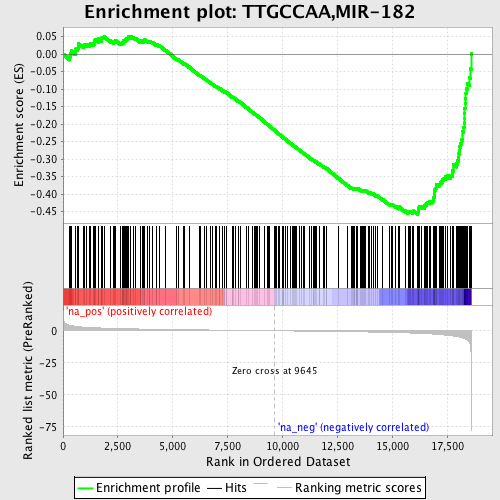
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTGCCAA,MIR-182 |
| Enrichment Score (ES) | -0.4581227 |
| Normalized Enrichment Score (NES) | -1.7343777 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09674076 |
| FWER p-Value | 0.78 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CFL1 | 309 | 4.276 | -0.0069 | No | ||
| 2 | NCALD | 339 | 4.155 | 0.0012 | No | ||
| 3 | PPIL1 | 362 | 4.062 | 0.0095 | No | ||
| 4 | BRE | 554 | 3.529 | 0.0073 | No | ||
| 5 | MTCH2 | 567 | 3.494 | 0.0147 | No | ||
| 6 | DENR | 673 | 3.237 | 0.0165 | No | ||
| 7 | PYGO2 | 690 | 3.192 | 0.0231 | No | ||
| 8 | RHOBTB1 | 705 | 3.164 | 0.0297 | No | ||
| 9 | RALGDS | 921 | 2.832 | 0.0246 | No | ||
| 10 | NFASC | 988 | 2.757 | 0.0274 | No | ||
| 11 | ARHGEF3 | 1081 | 2.668 | 0.0286 | No | ||
| 12 | P2RX4 | 1188 | 2.562 | 0.0288 | No | ||
| 13 | EIF3S10 | 1250 | 2.506 | 0.0313 | No | ||
| 14 | NAP1L1 | 1386 | 2.373 | 0.0295 | No | ||
| 15 | KTN1 | 1420 | 2.342 | 0.0331 | No | ||
| 16 | EPHA7 | 1427 | 2.337 | 0.0382 | No | ||
| 17 | MYOHD1 | 1470 | 2.304 | 0.0413 | No | ||
| 18 | DCUN1D1 | 1598 | 2.222 | 0.0395 | No | ||
| 19 | SEPT7 | 1620 | 2.211 | 0.0435 | No | ||
| 20 | RDX | 1751 | 2.130 | 0.0414 | No | ||
| 21 | SH3BP4 | 1757 | 2.124 | 0.0461 | No | ||
| 22 | GMFB | 1797 | 2.101 | 0.0488 | No | ||
| 23 | KDELR1 | 1869 | 2.058 | 0.0498 | No | ||
| 24 | ACACA | 2154 | 1.907 | 0.0387 | No | ||
| 25 | RGS17 | 2313 | 1.833 | 0.0344 | No | ||
| 26 | AUP1 | 2361 | 1.809 | 0.0361 | No | ||
| 27 | ARRDC3 | 2397 | 1.797 | 0.0383 | No | ||
| 28 | DAZAP2 | 2607 | 1.694 | 0.0309 | No | ||
| 29 | CORO1C | 2685 | 1.652 | 0.0306 | No | ||
| 30 | YAF2 | 2713 | 1.642 | 0.0329 | No | ||
| 31 | TEX2 | 2759 | 1.627 | 0.0342 | No | ||
| 32 | ATP2A2 | 2760 | 1.626 | 0.0380 | No | ||
| 33 | HOXA9 | 2809 | 1.606 | 0.0391 | No | ||
| 34 | ARMC1 | 2849 | 1.594 | 0.0407 | No | ||
| 35 | EGR3 | 2875 | 1.586 | 0.0431 | No | ||
| 36 | ZMPSTE24 | 2889 | 1.582 | 0.0460 | No | ||
| 37 | EVI5 | 2955 | 1.555 | 0.0461 | No | ||
| 38 | TXNL1 | 2964 | 1.552 | 0.0493 | No | ||
| 39 | PPP3R1 | 3054 | 1.521 | 0.0480 | No | ||
| 40 | EIF2C1 | 3056 | 1.521 | 0.0514 | No | ||
| 41 | UBE2R2 | 3206 | 1.466 | 0.0467 | No | ||
| 42 | EDNRB | 3286 | 1.437 | 0.0458 | No | ||
| 43 | CLOCK | 3519 | 1.357 | 0.0363 | No | ||
| 44 | DSCAM | 3531 | 1.354 | 0.0389 | No | ||
| 45 | LHX3 | 3603 | 1.335 | 0.0381 | No | ||
| 46 | NUMB | 3678 | 1.311 | 0.0371 | No | ||
| 47 | GABBR1 | 3693 | 1.306 | 0.0394 | No | ||
| 48 | HMGCLL1 | 3722 | 1.299 | 0.0409 | No | ||
| 49 | PPP1R2 | 3849 | 1.258 | 0.0370 | No | ||
| 50 | QKI | 3943 | 1.229 | 0.0348 | No | ||
| 51 | MBNL2 | 3950 | 1.227 | 0.0373 | No | ||
| 52 | EBF3 | 4056 | 1.198 | 0.0344 | No | ||
| 53 | CEBPA | 4256 | 1.143 | 0.0262 | No | ||
| 54 | SNX4 | 4270 | 1.140 | 0.0282 | No | ||
| 55 | BDNF | 4373 | 1.119 | 0.0252 | No | ||
| 56 | KPNA3 | 4672 | 1.044 | 0.0114 | No | ||
| 57 | NTNG1 | 5178 | 0.924 | -0.0139 | No | ||
| 58 | PRRX1 | 5245 | 0.907 | -0.0153 | No | ||
| 59 | PDZD4 | 5500 | 0.843 | -0.0272 | No | ||
| 60 | CTTN | 5551 | 0.833 | -0.0280 | No | ||
| 61 | RARG | 5744 | 0.786 | -0.0366 | No | ||
| 62 | SATB2 | 6211 | 0.688 | -0.0603 | No | ||
| 63 | FGF9 | 6249 | 0.678 | -0.0608 | No | ||
| 64 | CDV3 | 6442 | 0.638 | -0.0697 | No | ||
| 65 | BHLHB5 | 6545 | 0.620 | -0.0738 | No | ||
| 66 | SCRT1 | 6705 | 0.589 | -0.0811 | No | ||
| 67 | GPR85 | 6814 | 0.567 | -0.0857 | No | ||
| 68 | PCDH18 | 6950 | 0.541 | -0.0917 | No | ||
| 69 | LIMK1 | 6995 | 0.530 | -0.0929 | No | ||
| 70 | TSNAX | 7124 | 0.506 | -0.0987 | No | ||
| 71 | FZD3 | 7127 | 0.506 | -0.0976 | No | ||
| 72 | PCDH8 | 7275 | 0.470 | -0.1045 | No | ||
| 73 | VAMP3 | 7374 | 0.449 | -0.1088 | No | ||
| 74 | MMP16 | 7376 | 0.449 | -0.1078 | No | ||
| 75 | UBXD3 | 7440 | 0.437 | -0.1102 | No | ||
| 76 | ADAM10 | 7445 | 0.435 | -0.1094 | No | ||
| 77 | JUB | 7700 | 0.388 | -0.1223 | No | ||
| 78 | STARD13 | 7727 | 0.383 | -0.1229 | No | ||
| 79 | CHES1 | 7768 | 0.375 | -0.1242 | No | ||
| 80 | ATP1B3 | 7870 | 0.351 | -0.1288 | No | ||
| 81 | SEPT9 | 7978 | 0.330 | -0.1339 | No | ||
| 82 | TOPBP1 | 7998 | 0.326 | -0.1342 | No | ||
| 83 | BHLHB3 | 8100 | 0.303 | -0.1390 | No | ||
| 84 | ANUBL1 | 8358 | 0.253 | -0.1523 | No | ||
| 85 | RAC1 | 8465 | 0.231 | -0.1576 | No | ||
| 86 | ANK3 | 8646 | 0.196 | -0.1669 | No | ||
| 87 | NKX2-2 | 8721 | 0.180 | -0.1705 | No | ||
| 88 | EFNB2 | 8778 | 0.169 | -0.1732 | No | ||
| 89 | INSIG1 | 8825 | 0.159 | -0.1753 | No | ||
| 90 | THBS2 | 8865 | 0.151 | -0.1771 | No | ||
| 91 | PHF21A | 8950 | 0.135 | -0.1813 | No | ||
| 92 | PLD1 | 9163 | 0.095 | -0.1926 | No | ||
| 93 | EPAS1 | 9335 | 0.062 | -0.2018 | No | ||
| 94 | ADAMTS18 | 9362 | 0.057 | -0.2031 | No | ||
| 95 | BRMS1L | 9416 | 0.046 | -0.2058 | No | ||
| 96 | ACVR1 | 9653 | -0.003 | -0.2187 | No | ||
| 97 | EIF5 | 9700 | -0.012 | -0.2211 | No | ||
| 98 | PTHLH | 9734 | -0.017 | -0.2229 | No | ||
| 99 | MAK | 9836 | -0.036 | -0.2283 | No | ||
| 100 | SUHW2 | 9872 | -0.044 | -0.2301 | No | ||
| 101 | ARF4 | 9979 | -0.066 | -0.2357 | No | ||
| 102 | MTSS1 | 9994 | -0.070 | -0.2363 | No | ||
| 103 | BCL2L12 | 10039 | -0.077 | -0.2385 | No | ||
| 104 | YWHAG | 10062 | -0.083 | -0.2395 | No | ||
| 105 | IBRDC2 | 10145 | -0.101 | -0.2437 | No | ||
| 106 | CD2AP | 10217 | -0.113 | -0.2473 | No | ||
| 107 | BRUNOL6 | 10353 | -0.138 | -0.2544 | No | ||
| 108 | ELL2 | 10473 | -0.161 | -0.2604 | No | ||
| 109 | RASA1 | 10493 | -0.165 | -0.2611 | No | ||
| 110 | CCNJ | 10567 | -0.182 | -0.2646 | No | ||
| 111 | PAIP2 | 10595 | -0.189 | -0.2657 | No | ||
| 112 | RIMS3 | 10631 | -0.199 | -0.2671 | No | ||
| 113 | BCL2L13 | 10783 | -0.233 | -0.2748 | No | ||
| 114 | EPHB1 | 10857 | -0.246 | -0.2782 | No | ||
| 115 | EXOC4 | 10971 | -0.270 | -0.2837 | No | ||
| 116 | MMD | 10988 | -0.275 | -0.2839 | No | ||
| 117 | MEF2C | 11230 | -0.324 | -0.2963 | No | ||
| 118 | MYADM | 11303 | -0.340 | -0.2994 | No | ||
| 119 | SYPL1 | 11398 | -0.360 | -0.3037 | No | ||
| 120 | SP3 | 11425 | -0.367 | -0.3042 | No | ||
| 121 | ANK2 | 11438 | -0.370 | -0.3040 | No | ||
| 122 | FBXW11 | 11523 | -0.388 | -0.3077 | No | ||
| 123 | SLC1A2 | 11542 | -0.392 | -0.3078 | No | ||
| 124 | IGSF3 | 11669 | -0.420 | -0.3136 | No | ||
| 125 | APLN | 11705 | -0.428 | -0.3145 | No | ||
| 126 | KLF15 | 11849 | -0.462 | -0.3212 | No | ||
| 127 | LPHN2 | 11896 | -0.470 | -0.3227 | No | ||
| 128 | NPTX1 | 11994 | -0.488 | -0.3268 | No | ||
| 129 | ZCCHC14 | 12023 | -0.494 | -0.3272 | No | ||
| 130 | MARCKS | 12541 | -0.614 | -0.3538 | No | ||
| 131 | BNC2 | 12955 | -0.710 | -0.3747 | No | ||
| 132 | ADRA2C | 13129 | -0.755 | -0.3823 | No | ||
| 133 | SLC4A7 | 13174 | -0.768 | -0.3829 | No | ||
| 134 | TMEM47 | 13243 | -0.787 | -0.3848 | No | ||
| 135 | BAG4 | 13261 | -0.790 | -0.3839 | No | ||
| 136 | FOXF2 | 13300 | -0.801 | -0.3841 | No | ||
| 137 | SOX2 | 13350 | -0.812 | -0.3848 | No | ||
| 138 | GRINL1A | 13374 | -0.817 | -0.3842 | No | ||
| 139 | MITF | 13419 | -0.828 | -0.3847 | No | ||
| 140 | L1CAM | 13423 | -0.829 | -0.3829 | No | ||
| 141 | AKAP7 | 13557 | -0.862 | -0.3881 | No | ||
| 142 | FMR1 | 13617 | -0.878 | -0.3893 | No | ||
| 143 | PRDM1 | 13667 | -0.890 | -0.3899 | No | ||
| 144 | NPM1 | 13708 | -0.901 | -0.3900 | No | ||
| 145 | ABCC3 | 13754 | -0.914 | -0.3903 | No | ||
| 146 | ADCY2 | 13785 | -0.923 | -0.3898 | No | ||
| 147 | ATOH8 | 13934 | -0.970 | -0.3956 | No | ||
| 148 | RET | 13973 | -0.980 | -0.3954 | No | ||
| 149 | CUL5 | 14054 | -1.000 | -0.3974 | No | ||
| 150 | VLDLR | 14126 | -1.021 | -0.3989 | No | ||
| 151 | PRKACB | 14256 | -1.060 | -0.4034 | No | ||
| 152 | ELL | 14329 | -1.084 | -0.4048 | No | ||
| 153 | SLC39A9 | 14567 | -1.153 | -0.4150 | No | ||
| 154 | PRPF4B | 14864 | -1.262 | -0.4282 | No | ||
| 155 | ABLIM1 | 14957 | -1.298 | -0.4302 | No | ||
| 156 | MEF2D | 15003 | -1.317 | -0.4296 | No | ||
| 157 | DOK4 | 15160 | -1.385 | -0.4348 | No | ||
| 158 | HOXA10 | 15302 | -1.444 | -0.4391 | No | ||
| 159 | ISL1 | 15313 | -1.449 | -0.4363 | No | ||
| 160 | GRM5 | 15589 | -1.603 | -0.4475 | No | ||
| 161 | TAF15 | 15721 | -1.675 | -0.4508 | No | ||
| 162 | MARK3 | 15764 | -1.699 | -0.4491 | No | ||
| 163 | WSB1 | 15839 | -1.739 | -0.4491 | No | ||
| 164 | MFAP3 | 15939 | -1.799 | -0.4503 | No | ||
| 165 | SLITRK4 | 15975 | -1.828 | -0.4480 | No | ||
| 166 | KCMF1 | 16163 | -1.962 | -0.4536 | Yes | ||
| 167 | VASP | 16178 | -1.968 | -0.4498 | Yes | ||
| 168 | DR1 | 16199 | -1.983 | -0.4462 | Yes | ||
| 169 | ELMO1 | 16212 | -1.992 | -0.4423 | Yes | ||
| 170 | ARHGEF7 | 16218 | -1.995 | -0.4379 | Yes | ||
| 171 | XPR1 | 16247 | -2.017 | -0.4347 | Yes | ||
| 172 | RAB10 | 16340 | -2.094 | -0.4349 | Yes | ||
| 173 | ADD3 | 16459 | -2.185 | -0.4362 | Yes | ||
| 174 | PRKD1 | 16464 | -2.190 | -0.4313 | Yes | ||
| 175 | TRIM8 | 16532 | -2.239 | -0.4298 | Yes | ||
| 176 | ARHGDIA | 16577 | -2.282 | -0.4269 | Yes | ||
| 177 | EPM2A | 16598 | -2.299 | -0.4226 | Yes | ||
| 178 | VAV2 | 16677 | -2.369 | -0.4213 | Yes | ||
| 179 | CITED2 | 16757 | -2.461 | -0.4199 | Yes | ||
| 180 | PHF13 | 16858 | -2.565 | -0.4194 | Yes | ||
| 181 | FBXW7 | 16895 | -2.604 | -0.4153 | Yes | ||
| 182 | KHDRBS3 | 16897 | -2.605 | -0.4093 | Yes | ||
| 183 | TOB1 | 16926 | -2.641 | -0.4047 | Yes | ||
| 184 | PIGA | 16932 | -2.651 | -0.3988 | Yes | ||
| 185 | ADCY6 | 16938 | -2.656 | -0.3929 | Yes | ||
| 186 | RAB6B | 16945 | -2.664 | -0.3870 | Yes | ||
| 187 | OGT | 16987 | -2.708 | -0.3830 | Yes | ||
| 188 | PCMT1 | 17001 | -2.720 | -0.3774 | Yes | ||
| 189 | MAP1LC3B | 17027 | -2.762 | -0.3723 | Yes | ||
| 190 | SNAP23 | 17136 | -2.903 | -0.3714 | Yes | ||
| 191 | BTBD11 | 17178 | -2.964 | -0.3668 | Yes | ||
| 192 | ETS2 | 17249 | -3.075 | -0.3634 | Yes | ||
| 193 | RTN4 | 17290 | -3.130 | -0.3583 | Yes | ||
| 194 | WASF2 | 17355 | -3.238 | -0.3543 | Yes | ||
| 195 | SPRY4 | 17419 | -3.351 | -0.3499 | Yes | ||
| 196 | CD47 | 17520 | -3.553 | -0.3471 | Yes | ||
| 197 | XBP1 | 17652 | -3.801 | -0.3454 | Yes | ||
| 198 | AEBP2 | 17747 | -3.998 | -0.3412 | Yes | ||
| 199 | FEM1C | 17757 | -4.026 | -0.3324 | Yes | ||
| 200 | DOCK9 | 17791 | -4.078 | -0.3247 | Yes | ||
| 201 | MAGI1 | 17793 | -4.080 | -0.3153 | Yes | ||
| 202 | GIT2 | 17930 | -4.482 | -0.3122 | Yes | ||
| 203 | ATXN1 | 17981 | -4.640 | -0.3042 | Yes | ||
| 204 | ZNRF1 | 18026 | -4.772 | -0.2955 | Yes | ||
| 205 | IHPK1 | 18039 | -4.820 | -0.2849 | Yes | ||
| 206 | FLOT1 | 18055 | -4.861 | -0.2745 | Yes | ||
| 207 | KLF13 | 18072 | -4.938 | -0.2639 | Yes | ||
| 208 | SMAD1 | 18124 | -5.151 | -0.2547 | Yes | ||
| 209 | UBE3C | 18171 | -5.440 | -0.2445 | Yes | ||
| 210 | PAFAH1B1 | 18207 | -5.572 | -0.2335 | Yes | ||
| 211 | DYNC1LI2 | 18208 | -5.573 | -0.2205 | Yes | ||
| 212 | WDR40A | 18258 | -5.846 | -0.2096 | Yes | ||
| 213 | BCL11A | 18279 | -6.014 | -0.1967 | Yes | ||
| 214 | CSNK1E | 18290 | -6.103 | -0.1831 | Yes | ||
| 215 | KLF7 | 18294 | -6.123 | -0.1690 | Yes | ||
| 216 | DYRK1A | 18303 | -6.167 | -0.1551 | Yes | ||
| 217 | ZFP36L1 | 18330 | -6.331 | -0.1418 | Yes | ||
| 218 | SH2D1A | 18342 | -6.444 | -0.1275 | Yes | ||
| 219 | RNF44 | 18358 | -6.612 | -0.1129 | Yes | ||
| 220 | PBX2 | 18371 | -6.712 | -0.0980 | Yes | ||
| 221 | SS18L1 | 18420 | -7.212 | -0.0838 | Yes | ||
| 222 | BCL2 | 18530 | -9.542 | -0.0676 | Yes | ||
| 223 | ZFP36 | 18566 | -12.074 | -0.0414 | Yes | ||
| 224 | BACH2 | 18599 | -18.982 | 0.0009 | Yes |