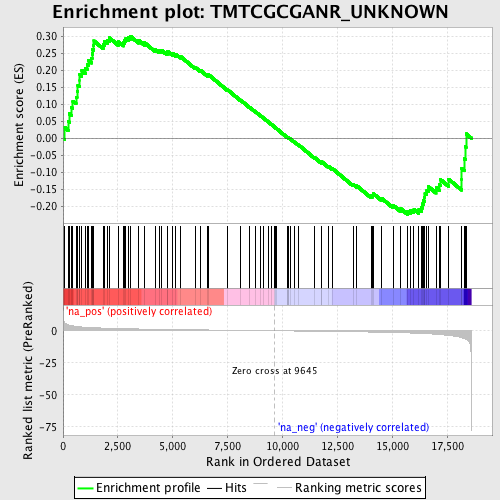
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TMTCGCGANR_UNKNOWN |
| Enrichment Score (ES) | 0.30010417 |
| Normalized Enrichment Score (NES) | 1.2456293 |
| Nominal p-value | 0.08713693 |
| FDR q-value | 0.5648167 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FUS | 68 | 6.156 | 0.0322 | Yes | ||
| 2 | PSMB2 | 231 | 4.585 | 0.0502 | Yes | ||
| 3 | POLDIP3 | 289 | 4.334 | 0.0724 | Yes | ||
| 4 | RPS19 | 387 | 3.975 | 0.0903 | Yes | ||
| 5 | SFRS5 | 446 | 3.781 | 0.1093 | Yes | ||
| 6 | RPS6 | 591 | 3.427 | 0.1215 | Yes | ||
| 7 | PRDX4 | 634 | 3.317 | 0.1385 | Yes | ||
| 8 | DENR | 673 | 3.237 | 0.1554 | Yes | ||
| 9 | SSR4 | 757 | 3.079 | 0.1688 | Yes | ||
| 10 | HSPA4 | 760 | 3.074 | 0.1867 | Yes | ||
| 11 | SCAMP5 | 854 | 2.926 | 0.1987 | Yes | ||
| 12 | MDH2 | 1021 | 2.727 | 0.2056 | Yes | ||
| 13 | RPS15A | 1095 | 2.653 | 0.2172 | Yes | ||
| 14 | SAP18 | 1144 | 2.598 | 0.2297 | Yes | ||
| 15 | RPL12 | 1304 | 2.447 | 0.2354 | Yes | ||
| 16 | TROAP | 1321 | 2.428 | 0.2487 | Yes | ||
| 17 | DDX3X | 1349 | 2.406 | 0.2613 | Yes | ||
| 18 | MATR3 | 1394 | 2.367 | 0.2727 | Yes | ||
| 19 | RPL10A | 1405 | 2.356 | 0.2859 | Yes | ||
| 20 | VDAC3 | 1826 | 2.080 | 0.2754 | Yes | ||
| 21 | CCT8 | 1878 | 2.050 | 0.2846 | Yes | ||
| 22 | CNOT3 | 2018 | 1.975 | 0.2886 | Yes | ||
| 23 | LUC7L2 | 2108 | 1.932 | 0.2950 | Yes | ||
| 24 | CDC5L | 2508 | 1.736 | 0.2836 | Yes | ||
| 25 | SLC25A4 | 2741 | 1.631 | 0.2806 | Yes | ||
| 26 | ATP5F1 | 2805 | 1.609 | 0.2866 | Yes | ||
| 27 | DCTN2 | 2860 | 1.590 | 0.2929 | Yes | ||
| 28 | ATF7 | 2981 | 1.544 | 0.2955 | Yes | ||
| 29 | DBR1 | 3060 | 1.519 | 0.3001 | Yes | ||
| 30 | ASH2L | 3439 | 1.385 | 0.2878 | No | ||
| 31 | SEC63 | 3713 | 1.301 | 0.2806 | No | ||
| 32 | SUV39H1 | 4210 | 1.155 | 0.2606 | No | ||
| 33 | UBE2D3 | 4386 | 1.114 | 0.2576 | No | ||
| 34 | BZW1 | 4489 | 1.088 | 0.2585 | No | ||
| 35 | PURA | 4742 | 1.026 | 0.2509 | No | ||
| 36 | FGFR1OP2 | 4765 | 1.021 | 0.2556 | No | ||
| 37 | MAP3K7 | 5000 | 0.970 | 0.2486 | No | ||
| 38 | COX7B | 5144 | 0.933 | 0.2464 | No | ||
| 39 | LRSAM1 | 5343 | 0.878 | 0.2408 | No | ||
| 40 | PRDX1 | 6011 | 0.730 | 0.2090 | No | ||
| 41 | TLOC1 | 6261 | 0.675 | 0.1995 | No | ||
| 42 | OFD1 | 6589 | 0.611 | 0.1854 | No | ||
| 43 | FSIP1 | 6614 | 0.606 | 0.1877 | No | ||
| 44 | NDUFA11 | 7499 | 0.425 | 0.1424 | No | ||
| 45 | IDH3G | 8083 | 0.307 | 0.1128 | No | ||
| 46 | BANP | 8513 | 0.222 | 0.0909 | No | ||
| 47 | PSMD5 | 8760 | 0.172 | 0.0786 | No | ||
| 48 | TAS1R1 | 8979 | 0.129 | 0.0676 | No | ||
| 49 | CHUK | 9136 | 0.102 | 0.0598 | No | ||
| 50 | PIGN | 9339 | 0.061 | 0.0492 | No | ||
| 51 | RPL26 | 9485 | 0.029 | 0.0416 | No | ||
| 52 | CAMK2D | 9652 | -0.002 | 0.0326 | No | ||
| 53 | EIF5 | 9700 | -0.012 | 0.0301 | No | ||
| 54 | KAZALD1 | 9706 | -0.012 | 0.0299 | No | ||
| 55 | CD2AP | 10217 | -0.113 | 0.0031 | No | ||
| 56 | RPS7 | 10247 | -0.120 | 0.0022 | No | ||
| 57 | PTPN4 | 10282 | -0.126 | 0.0011 | No | ||
| 58 | THAP7 | 10286 | -0.126 | 0.0017 | No | ||
| 59 | ANAPC4 | 10369 | -0.141 | -0.0019 | No | ||
| 60 | CCNJ | 10567 | -0.182 | -0.0115 | No | ||
| 61 | NUDT2 | 10739 | -0.222 | -0.0194 | No | ||
| 62 | TTC14 | 11471 | -0.379 | -0.0567 | No | ||
| 63 | WDR6 | 11773 | -0.445 | -0.0703 | No | ||
| 64 | UPF3B | 11783 | -0.447 | -0.0682 | No | ||
| 65 | COX11 | 12078 | -0.507 | -0.0811 | No | ||
| 66 | NFYC | 12267 | -0.548 | -0.0881 | No | ||
| 67 | RRAGA | 13214 | -0.780 | -0.1346 | No | ||
| 68 | RPL17 | 13372 | -0.816 | -0.1383 | No | ||
| 69 | POU4F1 | 14033 | -0.994 | -0.1682 | No | ||
| 70 | STXBP4 | 14105 | -1.015 | -0.1661 | No | ||
| 71 | PPP1R12A | 14148 | -1.028 | -0.1624 | No | ||
| 72 | ZZZ3 | 14501 | -1.135 | -0.1747 | No | ||
| 73 | GAS8 | 15039 | -1.334 | -0.1959 | No | ||
| 74 | MAPK14 | 15392 | -1.490 | -0.2063 | No | ||
| 75 | THAP1 | 15704 | -1.666 | -0.2133 | No | ||
| 76 | LAMP1 | 15829 | -1.733 | -0.2099 | No | ||
| 77 | MRP63 | 15990 | -1.839 | -0.2078 | No | ||
| 78 | SMARCD2 | 16216 | -1.994 | -0.2084 | No | ||
| 79 | MBTPS1 | 16344 | -2.096 | -0.2030 | No | ||
| 80 | BCAS3 | 16370 | -2.114 | -0.1920 | No | ||
| 81 | SLC12A2 | 16421 | -2.158 | -0.1821 | No | ||
| 82 | DGUOK | 16481 | -2.203 | -0.1725 | No | ||
| 83 | SUPT5H | 16493 | -2.215 | -0.1602 | No | ||
| 84 | PLOD2 | 16579 | -2.284 | -0.1514 | No | ||
| 85 | ATF2 | 16651 | -2.349 | -0.1416 | No | ||
| 86 | ARF3 | 16996 | -2.715 | -0.1443 | No | ||
| 87 | TLK2 | 17145 | -2.916 | -0.1353 | No | ||
| 88 | HNRPK | 17197 | -2.993 | -0.1206 | No | ||
| 89 | ANKFY1 | 17568 | -3.629 | -0.1194 | No | ||
| 90 | PFKFB2 | 18163 | -5.386 | -0.1201 | No | ||
| 91 | ZNF346 | 18169 | -5.435 | -0.0887 | No | ||
| 92 | UHRF2 | 18295 | -6.129 | -0.0597 | No | ||
| 93 | SLC25A11 | 18345 | -6.504 | -0.0244 | No | ||
| 94 | RAB33B | 18367 | -6.682 | 0.0134 | No |
