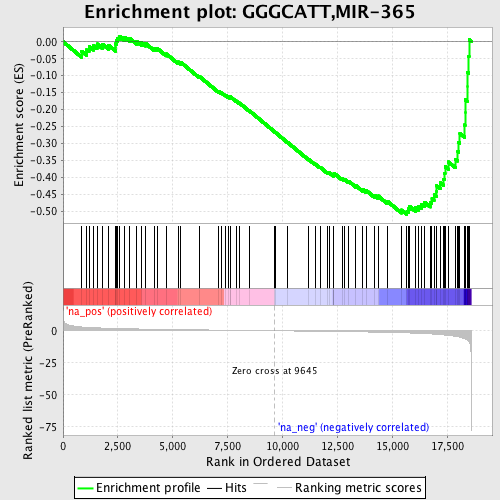
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GGGCATT,MIR-365 |
| Enrichment Score (ES) | -0.5086619 |
| Normalized Enrichment Score (NES) | -1.723135 |
| Nominal p-value | 0.0064683054 |
| FDR q-value | 0.09700797 |
| FWER p-Value | 0.814 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ING3 | 842 | 2.947 | -0.0278 | No | ||
| 2 | YTHDF2 | 1071 | 2.684 | -0.0240 | No | ||
| 3 | ANKRD17 | 1182 | 2.571 | -0.0146 | No | ||
| 4 | SOCS5 | 1369 | 2.387 | -0.0104 | No | ||
| 5 | SNTB2 | 1548 | 2.257 | -0.0065 | No | ||
| 6 | PPP5C | 1794 | 2.103 | -0.0071 | No | ||
| 7 | USP33 | 2074 | 1.950 | -0.0105 | No | ||
| 8 | PHF15 | 2394 | 1.798 | -0.0169 | No | ||
| 9 | ARRDC3 | 2397 | 1.797 | -0.0063 | No | ||
| 10 | DLX3 | 2423 | 1.782 | 0.0030 | No | ||
| 11 | GALNT4 | 2474 | 1.753 | 0.0108 | No | ||
| 12 | LIN7C | 2558 | 1.714 | 0.0166 | No | ||
| 13 | NR3C2 | 2795 | 1.613 | 0.0135 | No | ||
| 14 | DTNA | 3020 | 1.533 | 0.0106 | No | ||
| 15 | API5 | 3351 | 1.412 | 0.0012 | No | ||
| 16 | ARRDC2 | 3573 | 1.342 | -0.0027 | No | ||
| 17 | BMP1 | 3760 | 1.288 | -0.0050 | No | ||
| 18 | TRHDE | 4151 | 1.173 | -0.0190 | No | ||
| 19 | MAP2K7 | 4298 | 1.135 | -0.0201 | No | ||
| 20 | REV3L | 4704 | 1.036 | -0.0358 | No | ||
| 21 | COL7A1 | 5243 | 0.908 | -0.0594 | No | ||
| 22 | SIX4 | 5366 | 0.872 | -0.0607 | No | ||
| 23 | SATB2 | 6211 | 0.688 | -0.1022 | No | ||
| 24 | AMMECR1 | 7094 | 0.511 | -0.1467 | No | ||
| 25 | EFEMP1 | 7220 | 0.482 | -0.1506 | No | ||
| 26 | ADM | 7393 | 0.446 | -0.1572 | No | ||
| 27 | KLK15 | 7548 | 0.416 | -0.1630 | No | ||
| 28 | TMOD3 | 7607 | 0.404 | -0.1637 | No | ||
| 29 | ARHGAP12 | 7615 | 0.404 | -0.1617 | No | ||
| 30 | SLC8A2 | 7891 | 0.346 | -0.1744 | No | ||
| 31 | TLL2 | 8035 | 0.319 | -0.1802 | No | ||
| 32 | LAMP2 | 8493 | 0.225 | -0.2035 | No | ||
| 33 | ACVR1 | 9653 | -0.003 | -0.2661 | No | ||
| 34 | ARRB2 | 9665 | -0.005 | -0.2666 | No | ||
| 35 | RAPGEF4 | 10221 | -0.114 | -0.2959 | No | ||
| 36 | HMGCR | 11194 | -0.317 | -0.3464 | No | ||
| 37 | CREB5 | 11489 | -0.382 | -0.3600 | No | ||
| 38 | RBM12 | 11708 | -0.430 | -0.3692 | No | ||
| 39 | ST8SIA4 | 12053 | -0.501 | -0.3847 | No | ||
| 40 | SRGAP1 | 12128 | -0.519 | -0.3856 | No | ||
| 41 | SESTD1 | 12317 | -0.558 | -0.3924 | No | ||
| 42 | HDAC9 | 12339 | -0.563 | -0.3902 | No | ||
| 43 | FAM60A | 12344 | -0.565 | -0.3870 | No | ||
| 44 | PLCB4 | 12711 | -0.655 | -0.4029 | No | ||
| 45 | TIAM2 | 12838 | -0.682 | -0.4056 | No | ||
| 46 | NR4A2 | 13020 | -0.725 | -0.4110 | No | ||
| 47 | PAX6 | 13346 | -0.810 | -0.4237 | No | ||
| 48 | PRDM1 | 13667 | -0.890 | -0.4356 | No | ||
| 49 | PTGDR | 13825 | -0.936 | -0.4385 | No | ||
| 50 | PAX2 | 14212 | -1.050 | -0.4531 | No | ||
| 51 | ESRRA | 14352 | -1.089 | -0.4540 | No | ||
| 52 | CSK | 14774 | -1.225 | -0.4694 | No | ||
| 53 | ETV1 | 15424 | -1.513 | -0.4954 | No | ||
| 54 | HHIP | 15671 | -1.650 | -0.4988 | Yes | ||
| 55 | USP48 | 15734 | -1.681 | -0.4921 | Yes | ||
| 56 | MYLK | 15792 | -1.714 | -0.4849 | Yes | ||
| 57 | PCNP | 16055 | -1.878 | -0.4878 | Yes | ||
| 58 | ENTPD7 | 16209 | -1.989 | -0.4842 | Yes | ||
| 59 | SGK | 16324 | -2.082 | -0.4779 | Yes | ||
| 60 | ADD3 | 16459 | -2.185 | -0.4720 | Yes | ||
| 61 | VGLL4 | 16731 | -2.435 | -0.4721 | Yes | ||
| 62 | ETS1 | 16812 | -2.516 | -0.4614 | Yes | ||
| 63 | ARMCX3 | 16908 | -2.621 | -0.4508 | Yes | ||
| 64 | PKD2L2 | 17021 | -2.746 | -0.4404 | Yes | ||
| 65 | FMN2 | 17029 | -2.763 | -0.4243 | Yes | ||
| 66 | LDB1 | 17198 | -2.997 | -0.4154 | Yes | ||
| 67 | WDR37 | 17351 | -3.235 | -0.4043 | Yes | ||
| 68 | YWHAH | 17395 | -3.317 | -0.3868 | Yes | ||
| 69 | UBP1 | 17408 | -3.333 | -0.3675 | Yes | ||
| 70 | RAB22A | 17555 | -3.603 | -0.3538 | Yes | ||
| 71 | MYLIP | 17879 | -4.319 | -0.3454 | Yes | ||
| 72 | PHF12 | 17991 | -4.670 | -0.3235 | Yes | ||
| 73 | ZNRF1 | 18026 | -4.772 | -0.2967 | Yes | ||
| 74 | HIPK1 | 18085 | -4.982 | -0.2701 | Yes | ||
| 75 | BCL11B | 18306 | -6.176 | -0.2450 | Yes | ||
| 76 | SSH2 | 18321 | -6.266 | -0.2083 | Yes | ||
| 77 | SNRK | 18324 | -6.276 | -0.1709 | Yes | ||
| 78 | PLEKHA7 | 18417 | -7.197 | -0.1328 | Yes | ||
| 79 | AKT3 | 18419 | -7.198 | -0.0898 | Yes | ||
| 80 | SGK3 | 18490 | -8.275 | -0.0441 | Yes | ||
| 81 | KCNH2 | 18503 | -8.493 | 0.0061 | Yes |
