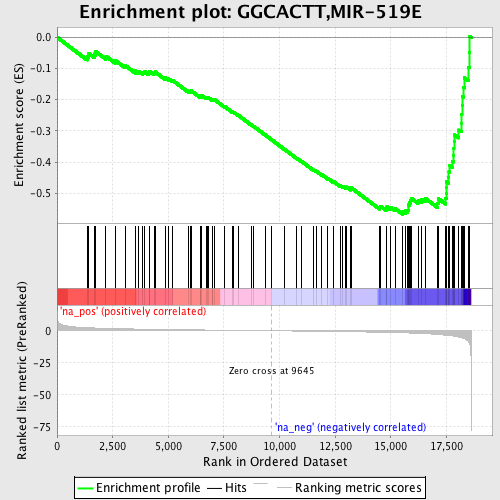
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GGCACTT,MIR-519E |
| Enrichment Score (ES) | -0.5676152 |
| Normalized Enrichment Score (NES) | -1.9645646 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.11103735 |
| FWER p-Value | 0.175 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDX3X | 1349 | 2.406 | -0.0601 | No | ||
| 2 | EPHA7 | 1427 | 2.337 | -0.0519 | No | ||
| 3 | RPS6KA3 | 1667 | 2.178 | -0.0533 | No | ||
| 4 | SEMA4G | 1738 | 2.136 | -0.0459 | No | ||
| 5 | SON | 2194 | 1.886 | -0.0605 | No | ||
| 6 | DAZAP2 | 2607 | 1.694 | -0.0738 | No | ||
| 7 | EIF2C1 | 3056 | 1.521 | -0.0899 | No | ||
| 8 | CLOCK | 3519 | 1.357 | -0.1077 | No | ||
| 9 | ZNF148 | 3661 | 1.314 | -0.1084 | No | ||
| 10 | IRF1 | 3854 | 1.258 | -0.1121 | No | ||
| 11 | QKI | 3943 | 1.229 | -0.1103 | No | ||
| 12 | C1GALT1 | 4130 | 1.177 | -0.1142 | No | ||
| 13 | NPAS2 | 4168 | 1.168 | -0.1100 | No | ||
| 14 | VPS26A | 4357 | 1.123 | -0.1142 | No | ||
| 15 | MMP19 | 4404 | 1.109 | -0.1109 | No | ||
| 16 | MYST3 | 4851 | 1.002 | -0.1296 | No | ||
| 17 | MAP3K7 | 5000 | 0.970 | -0.1325 | No | ||
| 18 | CCNT2 | 5179 | 0.924 | -0.1372 | No | ||
| 19 | MTMR4 | 5897 | 0.755 | -0.1720 | No | ||
| 20 | LBP | 6013 | 0.730 | -0.1743 | No | ||
| 21 | ARHGEF12 | 6020 | 0.728 | -0.1708 | No | ||
| 22 | DDX3Y | 6443 | 0.638 | -0.1902 | No | ||
| 23 | CORO7 | 6444 | 0.638 | -0.1868 | No | ||
| 24 | ARID4A | 6489 | 0.631 | -0.1859 | No | ||
| 25 | DKK1 | 6695 | 0.591 | -0.1938 | No | ||
| 26 | PAFAH1B2 | 6757 | 0.579 | -0.1941 | No | ||
| 27 | PTPN3 | 6787 | 0.573 | -0.1926 | No | ||
| 28 | ZBTB7A | 6998 | 0.530 | -0.2012 | No | ||
| 29 | NR2F2 | 7002 | 0.528 | -0.1985 | No | ||
| 30 | DPYSL5 | 7068 | 0.517 | -0.1993 | No | ||
| 31 | ST8SIA2 | 7527 | 0.418 | -0.2218 | No | ||
| 32 | EFNB3 | 7863 | 0.352 | -0.2381 | No | ||
| 33 | EIF5A2 | 7922 | 0.341 | -0.2394 | No | ||
| 34 | PTGFRN | 8147 | 0.294 | -0.2499 | No | ||
| 35 | ZBTB4 | 8728 | 0.179 | -0.2803 | No | ||
| 36 | FGD1 | 8827 | 0.159 | -0.2847 | No | ||
| 37 | SCML2 | 9353 | 0.059 | -0.3128 | No | ||
| 38 | SOX11 | 9645 | -0.000 | -0.3285 | No | ||
| 39 | PCYT1B | 10236 | -0.118 | -0.3597 | No | ||
| 40 | BTBD7 | 10772 | -0.231 | -0.3874 | No | ||
| 41 | MMD | 10988 | -0.275 | -0.3975 | No | ||
| 42 | SSFA2 | 11535 | -0.391 | -0.4249 | No | ||
| 43 | ZFHX4 | 11646 | -0.415 | -0.4287 | No | ||
| 44 | LPHN2 | 11896 | -0.470 | -0.4396 | No | ||
| 45 | EDG1 | 12175 | -0.527 | -0.4519 | No | ||
| 46 | PTPRD | 12436 | -0.588 | -0.4628 | No | ||
| 47 | TBL1X | 12729 | -0.658 | -0.4751 | No | ||
| 48 | DLL1 | 12845 | -0.683 | -0.4777 | No | ||
| 49 | XRN1 | 12967 | -0.713 | -0.4805 | No | ||
| 50 | NR4A2 | 13020 | -0.725 | -0.4794 | No | ||
| 51 | MGEA5 | 13206 | -0.777 | -0.4853 | No | ||
| 52 | PHF1 | 13216 | -0.780 | -0.4817 | No | ||
| 53 | NANOS1 | 14488 | -1.132 | -0.5443 | No | ||
| 54 | WAC | 14532 | -1.143 | -0.5406 | No | ||
| 55 | PTPRG | 14787 | -1.230 | -0.5478 | No | ||
| 56 | FURIN | 14825 | -1.246 | -0.5433 | No | ||
| 57 | MARK4 | 15005 | -1.319 | -0.5460 | No | ||
| 58 | PTEN | 15194 | -1.398 | -0.5487 | No | ||
| 59 | CDK10 | 15545 | -1.583 | -0.5593 | Yes | ||
| 60 | TAGAP | 15650 | -1.638 | -0.5562 | Yes | ||
| 61 | SP1 | 15765 | -1.699 | -0.5534 | Yes | ||
| 62 | MYLK | 15792 | -1.714 | -0.5458 | Yes | ||
| 63 | RBL2 | 15793 | -1.714 | -0.5367 | Yes | ||
| 64 | BCOR | 15828 | -1.731 | -0.5294 | Yes | ||
| 65 | BNIP2 | 15896 | -1.770 | -0.5237 | Yes | ||
| 66 | OSR1 | 15907 | -1.777 | -0.5148 | Yes | ||
| 67 | AOF1 | 16222 | -2.000 | -0.5212 | Yes | ||
| 68 | RRM2 | 16378 | -2.120 | -0.5184 | Yes | ||
| 69 | ZNF385 | 16544 | -2.248 | -0.5155 | Yes | ||
| 70 | SPATA2 | 17111 | -2.869 | -0.5309 | Yes | ||
| 71 | BICD2 | 17140 | -2.908 | -0.5170 | Yes | ||
| 72 | PPP2R2A | 17459 | -3.424 | -0.5161 | Yes | ||
| 73 | RAB35 | 17493 | -3.492 | -0.4995 | Yes | ||
| 74 | PERQ1 | 17499 | -3.504 | -0.4812 | Yes | ||
| 75 | CXCR4 | 17506 | -3.516 | -0.4630 | Yes | ||
| 76 | ZIC4 | 17592 | -3.667 | -0.4482 | Yes | ||
| 77 | SERTAD2 | 17612 | -3.708 | -0.4297 | Yes | ||
| 78 | MAT2B | 17651 | -3.800 | -0.4117 | Yes | ||
| 79 | MKRN1 | 17773 | -4.056 | -0.3968 | Yes | ||
| 80 | FBXO21 | 17826 | -4.170 | -0.3776 | Yes | ||
| 81 | PLAGL2 | 17833 | -4.192 | -0.3558 | Yes | ||
| 82 | ARID4B | 17840 | -4.206 | -0.3339 | Yes | ||
| 83 | SRPK1 | 17868 | -4.293 | -0.3127 | Yes | ||
| 84 | CCNG2 | 18053 | -4.860 | -0.2970 | Yes | ||
| 85 | UBE3C | 18171 | -5.440 | -0.2746 | Yes | ||
| 86 | RAB5B | 18176 | -5.449 | -0.2460 | Yes | ||
| 87 | MARK2 | 18200 | -5.546 | -0.2180 | Yes | ||
| 88 | PAFAH1B1 | 18207 | -5.572 | -0.1889 | Yes | ||
| 89 | NEDD4L | 18277 | -5.997 | -0.1610 | Yes | ||
| 90 | BCL11B | 18306 | -6.176 | -0.1299 | Yes | ||
| 91 | ARHGEF5 | 18487 | -8.231 | -0.0962 | Yes | ||
| 92 | EHD3 | 18522 | -9.319 | -0.0488 | Yes | ||
| 93 | ARL4C | 18547 | -10.194 | 0.0037 | Yes |
