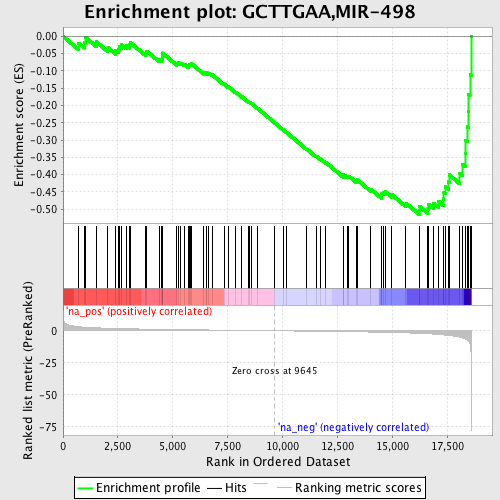
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GCTTGAA,MIR-498 |
| Enrichment Score (ES) | -0.5156312 |
| Normalized Enrichment Score (NES) | -1.7398523 |
| Nominal p-value | 0.004178273 |
| FDR q-value | 0.09765898 |
| FWER p-Value | 0.763 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NARG1 | 710 | 3.156 | -0.0193 | No | ||
| 2 | TFRC | 975 | 2.772 | -0.0169 | No | ||
| 3 | RNF125 | 1022 | 2.727 | -0.0029 | No | ||
| 4 | RUNX2 | 1507 | 2.283 | -0.0153 | No | ||
| 5 | FUSIP1 | 2044 | 1.962 | -0.0324 | No | ||
| 6 | ARRDC3 | 2397 | 1.797 | -0.0406 | No | ||
| 7 | DUSP4 | 2526 | 1.728 | -0.0371 | No | ||
| 8 | LRP1B | 2573 | 1.709 | -0.0293 | No | ||
| 9 | PALM2 | 2669 | 1.661 | -0.0244 | No | ||
| 10 | UBE2J1 | 2886 | 1.583 | -0.0265 | No | ||
| 11 | DTNA | 3020 | 1.533 | -0.0245 | No | ||
| 12 | PHF6 | 3072 | 1.514 | -0.0181 | No | ||
| 13 | CACNA2D3 | 3773 | 1.284 | -0.0482 | No | ||
| 14 | EXOC5 | 3819 | 1.271 | -0.0429 | No | ||
| 15 | KCND2 | 4374 | 1.118 | -0.0661 | No | ||
| 16 | CAPZA2 | 4487 | 1.089 | -0.0656 | No | ||
| 17 | EML4 | 4511 | 1.083 | -0.0603 | No | ||
| 18 | PDIA6 | 4522 | 1.080 | -0.0543 | No | ||
| 19 | WT1 | 4525 | 1.080 | -0.0480 | No | ||
| 20 | CCNT2 | 5179 | 0.924 | -0.0776 | No | ||
| 21 | RNF11 | 5236 | 0.908 | -0.0752 | No | ||
| 22 | SIX4 | 5366 | 0.872 | -0.0769 | No | ||
| 23 | PLEKHC1 | 5528 | 0.837 | -0.0805 | No | ||
| 24 | CBX4 | 5706 | 0.798 | -0.0853 | No | ||
| 25 | SOCS3 | 5739 | 0.788 | -0.0823 | No | ||
| 26 | MORF4L2 | 5791 | 0.776 | -0.0803 | No | ||
| 27 | PIK3R1 | 5830 | 0.769 | -0.0778 | No | ||
| 28 | DHX35 | 6410 | 0.646 | -0.1051 | No | ||
| 29 | CAMK2G | 6527 | 0.623 | -0.1076 | No | ||
| 30 | KPNB1 | 6534 | 0.622 | -0.1042 | No | ||
| 31 | PKNOX2 | 6641 | 0.600 | -0.1063 | No | ||
| 32 | COL1A1 | 6788 | 0.572 | -0.1107 | No | ||
| 33 | MAB21L1 | 7340 | 0.456 | -0.1377 | No | ||
| 34 | ST8SIA2 | 7527 | 0.418 | -0.1452 | No | ||
| 35 | OLIG3 | 7861 | 0.353 | -0.1611 | No | ||
| 36 | PACSIN2 | 8149 | 0.294 | -0.1748 | No | ||
| 37 | CPNE8 | 8471 | 0.230 | -0.1907 | No | ||
| 38 | ITSN1 | 8488 | 0.226 | -0.1902 | No | ||
| 39 | FLRT2 | 8566 | 0.212 | -0.1931 | No | ||
| 40 | RBPMS | 8867 | 0.151 | -0.2084 | No | ||
| 41 | SOX11 | 9645 | -0.000 | -0.2503 | No | ||
| 42 | DCBLD2 | 10033 | -0.076 | -0.2707 | No | ||
| 43 | NAP1L3 | 10172 | -0.104 | -0.2775 | No | ||
| 44 | CRH | 11111 | -0.301 | -0.3263 | No | ||
| 45 | SSFA2 | 11535 | -0.391 | -0.3468 | No | ||
| 46 | PAM | 11730 | -0.437 | -0.3546 | No | ||
| 47 | ATPAF1 | 11972 | -0.482 | -0.3647 | No | ||
| 48 | GRIA2 | 12768 | -0.666 | -0.4036 | No | ||
| 49 | RAB11A | 12786 | -0.669 | -0.4005 | No | ||
| 50 | MAPRE1 | 12962 | -0.712 | -0.4056 | No | ||
| 51 | CCNE1 | 13017 | -0.724 | -0.4042 | No | ||
| 52 | CHODL | 13387 | -0.821 | -0.4192 | No | ||
| 53 | FANCA | 13412 | -0.827 | -0.4155 | No | ||
| 54 | SPRED1 | 14022 | -0.992 | -0.4424 | No | ||
| 55 | RAB6A | 14514 | -1.140 | -0.4620 | No | ||
| 56 | DICER1 | 14530 | -1.143 | -0.4559 | No | ||
| 57 | PPP3CA | 14583 | -1.160 | -0.4517 | No | ||
| 58 | GJA1 | 14671 | -1.189 | -0.4493 | No | ||
| 59 | PDE4B | 14969 | -1.302 | -0.4575 | No | ||
| 60 | WDR26 | 15626 | -1.622 | -0.4831 | No | ||
| 61 | KLF12 | 16230 | -2.003 | -0.5036 | Yes | ||
| 62 | MEIS1 | 16251 | -2.021 | -0.4925 | Yes | ||
| 63 | RNF149 | 16628 | -2.325 | -0.4988 | Yes | ||
| 64 | MECP2 | 16664 | -2.359 | -0.4865 | Yes | ||
| 65 | ABTB1 | 16880 | -2.591 | -0.4825 | Yes | ||
| 66 | YPEL2 | 17115 | -2.872 | -0.4778 | Yes | ||
| 67 | IRF2 | 17342 | -3.221 | -0.4706 | Yes | ||
| 68 | CTNS | 17357 | -3.240 | -0.4519 | Yes | ||
| 69 | NCOA1 | 17413 | -3.345 | -0.4347 | Yes | ||
| 70 | RAB22A | 17555 | -3.603 | -0.4206 | Yes | ||
| 71 | ZIC4 | 17592 | -3.667 | -0.4005 | Yes | ||
| 72 | HIPK1 | 18085 | -4.982 | -0.3971 | Yes | ||
| 73 | CUGBP1 | 18217 | -5.616 | -0.3703 | Yes | ||
| 74 | SNRK | 18324 | -6.276 | -0.3383 | Yes | ||
| 75 | ZFP36L1 | 18330 | -6.331 | -0.3004 | Yes | ||
| 76 | SS18L1 | 18420 | -7.212 | -0.2618 | Yes | ||
| 77 | HBP1 | 18469 | -7.955 | -0.2165 | Yes | ||
| 78 | PURG | 18483 | -8.094 | -0.1685 | Yes | ||
| 79 | ARL4C | 18547 | -10.194 | -0.1106 | Yes | ||
| 80 | BACH2 | 18599 | -18.982 | 0.0009 | Yes |
