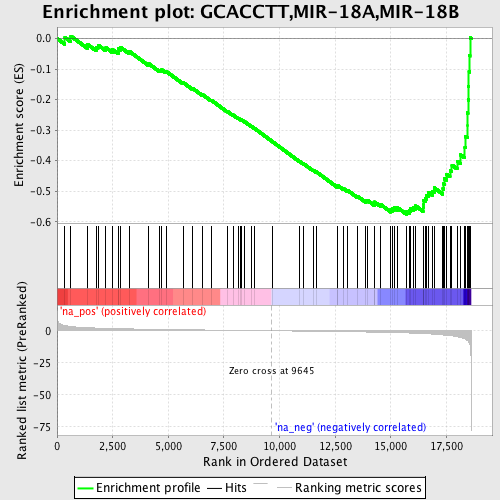
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GCACCTT,MIR-18A,MIR-18B |
| Enrichment Score (ES) | -0.5760494 |
| Normalized Enrichment Score (NES) | -1.9109559 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.073259935 |
| FWER p-Value | 0.271 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TRIM2 | 350 | 4.104 | 0.0038 | No | ||
| 2 | PHF19 | 618 | 3.359 | 0.0080 | No | ||
| 3 | SOCS5 | 1369 | 2.387 | -0.0192 | No | ||
| 4 | SH3BP4 | 1757 | 2.124 | -0.0284 | No | ||
| 5 | AKR1D1 | 1838 | 2.076 | -0.0212 | No | ||
| 6 | SON | 2194 | 1.886 | -0.0299 | No | ||
| 7 | IGF1 | 2496 | 1.741 | -0.0365 | No | ||
| 8 | CDC42 | 2751 | 1.629 | -0.0412 | No | ||
| 9 | TEX2 | 2759 | 1.627 | -0.0326 | No | ||
| 10 | EHMT1 | 2859 | 1.590 | -0.0291 | No | ||
| 11 | NR3C1 | 3247 | 1.453 | -0.0420 | No | ||
| 12 | PIAS3 | 4102 | 1.186 | -0.0815 | No | ||
| 13 | RAB11FIP2 | 4616 | 1.058 | -0.1033 | No | ||
| 14 | PRICKLE2 | 4708 | 1.036 | -0.1025 | No | ||
| 15 | MESP1 | 4898 | 0.992 | -0.1072 | No | ||
| 16 | DPP10 | 5670 | 0.806 | -0.1443 | No | ||
| 17 | HSF2 | 6089 | 0.713 | -0.1629 | No | ||
| 18 | BHLHB5 | 6545 | 0.620 | -0.1840 | No | ||
| 19 | ARL15 | 6920 | 0.544 | -0.2012 | No | ||
| 20 | ASXL2 | 7654 | 0.396 | -0.2385 | No | ||
| 21 | BTG3 | 7906 | 0.343 | -0.2502 | No | ||
| 22 | PTGFRN | 8147 | 0.294 | -0.2615 | No | ||
| 23 | LIN28 | 8250 | 0.271 | -0.2655 | No | ||
| 24 | CTGF | 8285 | 0.264 | -0.2659 | No | ||
| 25 | ALCAM | 8429 | 0.242 | -0.2722 | No | ||
| 26 | ZBTB4 | 8728 | 0.179 | -0.2873 | No | ||
| 27 | RABGAP1 | 8866 | 0.151 | -0.2939 | No | ||
| 28 | SIM2 | 9673 | -0.007 | -0.3373 | No | ||
| 29 | TARDBP | 10890 | -0.255 | -0.4015 | No | ||
| 30 | HIF1A | 11096 | -0.298 | -0.4109 | No | ||
| 31 | RHOT1 | 11518 | -0.388 | -0.4315 | No | ||
| 32 | CTDSPL | 11650 | -0.416 | -0.4362 | No | ||
| 33 | PURB | 12592 | -0.627 | -0.4835 | No | ||
| 34 | GCLC | 12623 | -0.633 | -0.4816 | No | ||
| 35 | NAV1 | 12858 | -0.687 | -0.4904 | No | ||
| 36 | KIT | 13043 | -0.731 | -0.4963 | No | ||
| 37 | SAR1A | 13496 | -0.847 | -0.5160 | No | ||
| 38 | TRIOBP | 13873 | -0.951 | -0.5310 | No | ||
| 39 | MAN1A2 | 13951 | -0.973 | -0.5298 | No | ||
| 40 | PRKACB | 14256 | -1.060 | -0.5403 | No | ||
| 41 | FRMD4A | 14268 | -1.067 | -0.5350 | No | ||
| 42 | ESR1 | 14537 | -1.146 | -0.5431 | No | ||
| 43 | MEF2D | 15003 | -1.317 | -0.5609 | No | ||
| 44 | GLRB | 15080 | -1.350 | -0.5576 | No | ||
| 45 | NR1H2 | 15158 | -1.384 | -0.5541 | No | ||
| 46 | CLASP2 | 15306 | -1.446 | -0.5540 | No | ||
| 47 | MDGA1 | 15716 | -1.672 | -0.5668 | Yes | ||
| 48 | WSB1 | 15839 | -1.739 | -0.5638 | Yes | ||
| 49 | FNBP1 | 15877 | -1.761 | -0.5560 | Yes | ||
| 50 | FCHSD2 | 16009 | -1.849 | -0.5529 | Yes | ||
| 51 | SMAD2 | 16115 | -1.921 | -0.5479 | Yes | ||
| 52 | XYLT2 | 16451 | -2.181 | -0.5539 | Yes | ||
| 53 | ADD3 | 16459 | -2.185 | -0.5422 | Yes | ||
| 54 | RUNX1 | 16466 | -2.190 | -0.5304 | Yes | ||
| 55 | CRIM1 | 16549 | -2.253 | -0.5224 | Yes | ||
| 56 | NFAT5 | 16614 | -2.316 | -0.5130 | Yes | ||
| 57 | TRIB2 | 16696 | -2.391 | -0.5042 | Yes | ||
| 58 | STK4 | 16872 | -2.580 | -0.4993 | Yes | ||
| 59 | ETV6 | 16949 | -2.672 | -0.4886 | Yes | ||
| 60 | IRF2 | 17342 | -3.221 | -0.4920 | Yes | ||
| 61 | RAB5C | 17378 | -3.276 | -0.4757 | Yes | ||
| 62 | NCOA1 | 17413 | -3.345 | -0.4591 | Yes | ||
| 63 | PERQ1 | 17499 | -3.504 | -0.4443 | Yes | ||
| 64 | CLK2 | 17661 | -3.812 | -0.4319 | Yes | ||
| 65 | AEBP2 | 17747 | -3.998 | -0.4144 | Yes | ||
| 66 | ATXN1 | 17981 | -4.640 | -0.4013 | Yes | ||
| 67 | JARID1B | 18138 | -5.235 | -0.3807 | Yes | ||
| 68 | ZFP36L1 | 18330 | -6.331 | -0.3560 | Yes | ||
| 69 | PARP6 | 18348 | -6.525 | -0.3208 | Yes | ||
| 70 | ANKRD50 | 18437 | -7.443 | -0.2844 | Yes | ||
| 71 | PACSIN1 | 18440 | -7.482 | -0.2431 | Yes | ||
| 72 | NEDD9 | 18470 | -7.962 | -0.2007 | Yes | ||
| 73 | ARHGEF5 | 18487 | -8.231 | -0.1560 | Yes | ||
| 74 | SNURF | 18512 | -8.802 | -0.1086 | Yes | ||
| 75 | PHF2 | 18536 | -9.766 | -0.0559 | Yes | ||
| 76 | GAB1 | 18560 | -10.871 | 0.0030 | Yes |
