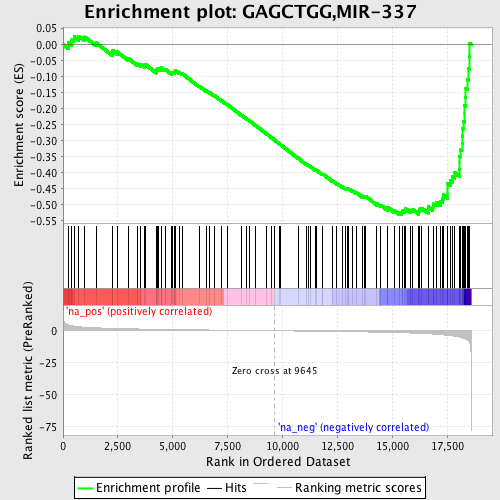
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GAGCTGG,MIR-337 |
| Enrichment Score (ES) | -0.5300276 |
| Normalized Enrichment Score (NES) | -1.8433484 |
| Nominal p-value | 0.0025542784 |
| FDR q-value | 0.084406696 |
| FWER p-Value | 0.434 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EI24 | 242 | 4.531 | 0.0064 | No | ||
| 2 | BCAP29 | 396 | 3.933 | 0.0150 | No | ||
| 3 | ABCF1 | 497 | 3.659 | 0.0253 | No | ||
| 4 | PDGFRB | 718 | 3.142 | 0.0269 | No | ||
| 5 | NFASC | 988 | 2.757 | 0.0242 | No | ||
| 6 | THRA | 1503 | 2.284 | 0.0062 | No | ||
| 7 | MAMDC1 | 2254 | 1.859 | -0.0263 | No | ||
| 8 | LRRC4 | 2258 | 1.858 | -0.0185 | No | ||
| 9 | IER5L | 2464 | 1.757 | -0.0220 | No | ||
| 10 | MFN1 | 2991 | 1.541 | -0.0438 | No | ||
| 11 | MCRS1 | 3377 | 1.402 | -0.0586 | No | ||
| 12 | DSCAM | 3531 | 1.354 | -0.0611 | No | ||
| 13 | NR1D1 | 3720 | 1.299 | -0.0656 | No | ||
| 14 | ACOT7 | 3766 | 1.285 | -0.0625 | No | ||
| 15 | EHMT2 | 4244 | 1.147 | -0.0834 | No | ||
| 16 | BNIPL | 4249 | 1.145 | -0.0787 | No | ||
| 17 | DPH2 | 4283 | 1.138 | -0.0756 | No | ||
| 18 | CD44 | 4364 | 1.122 | -0.0751 | No | ||
| 19 | H1F0 | 4482 | 1.090 | -0.0767 | No | ||
| 20 | LBX1 | 4485 | 1.089 | -0.0722 | No | ||
| 21 | RNF121 | 4660 | 1.048 | -0.0771 | No | ||
| 22 | KCNH8 | 4936 | 0.983 | -0.0877 | No | ||
| 23 | SEMA6A | 4968 | 0.977 | -0.0852 | No | ||
| 24 | TOR2A | 5069 | 0.954 | -0.0865 | No | ||
| 25 | GPD1 | 5071 | 0.954 | -0.0825 | No | ||
| 26 | KLF11 | 5110 | 0.943 | -0.0805 | No | ||
| 27 | SLC12A5 | 5287 | 0.895 | -0.0861 | No | ||
| 28 | TWIST1 | 5426 | 0.858 | -0.0899 | No | ||
| 29 | RAB15 | 6207 | 0.689 | -0.1291 | No | ||
| 30 | CAMK2G | 6527 | 0.623 | -0.1436 | No | ||
| 31 | RNF19 | 6686 | 0.593 | -0.1496 | No | ||
| 32 | MAP3K9 | 6919 | 0.544 | -0.1598 | No | ||
| 33 | MRPS16 | 7217 | 0.483 | -0.1738 | No | ||
| 34 | B3GAT3 | 7496 | 0.426 | -0.1870 | No | ||
| 35 | PACSIN2 | 8149 | 0.294 | -0.2210 | No | ||
| 36 | ABCA9 | 8359 | 0.252 | -0.2312 | No | ||
| 37 | TRIM35 | 8511 | 0.222 | -0.2384 | No | ||
| 38 | CHRD | 8764 | 0.171 | -0.2513 | No | ||
| 39 | VCP | 9269 | 0.078 | -0.2781 | No | ||
| 40 | RASGRP4 | 9480 | 0.031 | -0.2894 | No | ||
| 41 | COMMD3 | 9620 | 0.005 | -0.2968 | No | ||
| 42 | EFNB1 | 9863 | -0.041 | -0.3097 | No | ||
| 43 | KCND1 | 9894 | -0.048 | -0.3112 | No | ||
| 44 | ITGA11 | 10731 | -0.221 | -0.3554 | No | ||
| 45 | SP7 | 11098 | -0.298 | -0.3739 | No | ||
| 46 | SFRS1 | 11161 | -0.311 | -0.3759 | No | ||
| 47 | STIP1 | 11276 | -0.334 | -0.3806 | No | ||
| 48 | TYSND1 | 11498 | -0.384 | -0.3909 | No | ||
| 49 | SLC1A2 | 11542 | -0.392 | -0.3915 | No | ||
| 50 | FOS | 11810 | -0.455 | -0.4040 | No | ||
| 51 | AP1S3 | 11842 | -0.461 | -0.4037 | No | ||
| 52 | HOXB5 | 12281 | -0.553 | -0.4250 | No | ||
| 53 | FOSL1 | 12479 | -0.600 | -0.4331 | No | ||
| 54 | SLC25A22 | 12726 | -0.658 | -0.4435 | No | ||
| 55 | NAV1 | 12858 | -0.687 | -0.4477 | No | ||
| 56 | BNC2 | 12955 | -0.710 | -0.4498 | No | ||
| 57 | OVOL1 | 13021 | -0.725 | -0.4502 | No | ||
| 58 | TEAD3 | 13199 | -0.774 | -0.4564 | No | ||
| 59 | CHD4 | 13352 | -0.812 | -0.4612 | No | ||
| 60 | FRAG1 | 13629 | -0.880 | -0.4723 | No | ||
| 61 | MAT2A | 13726 | -0.905 | -0.4736 | No | ||
| 62 | JPH1 | 13790 | -0.924 | -0.4730 | No | ||
| 63 | GRK6 | 14305 | -1.077 | -0.4962 | No | ||
| 64 | ALDH1A2 | 14476 | -1.127 | -0.5005 | No | ||
| 65 | PPP1R3F | 14771 | -1.224 | -0.5112 | No | ||
| 66 | SSBP3 | 14795 | -1.235 | -0.5071 | No | ||
| 67 | SEC24C | 15086 | -1.353 | -0.5170 | No | ||
| 68 | ROM1 | 15310 | -1.448 | -0.5228 | No | ||
| 69 | B4GALT3 | 15445 | -1.526 | -0.5235 | Yes | ||
| 70 | TOMM34 | 15458 | -1.530 | -0.5176 | Yes | ||
| 71 | PARVA | 15565 | -1.593 | -0.5164 | Yes | ||
| 72 | FBXL17 | 15623 | -1.619 | -0.5126 | Yes | ||
| 73 | DGKZ | 15838 | -1.738 | -0.5167 | Yes | ||
| 74 | YPEL1 | 15928 | -1.791 | -0.5138 | Yes | ||
| 75 | SMARCD2 | 16216 | -1.994 | -0.5207 | Yes | ||
| 76 | KLF12 | 16230 | -2.003 | -0.5128 | Yes | ||
| 77 | ZMYND11 | 16341 | -2.094 | -0.5098 | Yes | ||
| 78 | SBK1 | 16633 | -2.330 | -0.5155 | Yes | ||
| 79 | LIMK2 | 16635 | -2.331 | -0.5056 | Yes | ||
| 80 | PHF13 | 16858 | -2.565 | -0.5066 | Yes | ||
| 81 | POGZ | 16891 | -2.601 | -0.4971 | Yes | ||
| 82 | SEPT6 | 17004 | -2.726 | -0.4915 | Yes | ||
| 83 | DNAJB5 | 17190 | -2.983 | -0.4887 | Yes | ||
| 84 | BTG2 | 17284 | -3.125 | -0.4803 | Yes | ||
| 85 | JUNB | 17330 | -3.198 | -0.4690 | Yes | ||
| 86 | BCL2L1 | 17522 | -3.558 | -0.4640 | Yes | ||
| 87 | TXNL4B | 17524 | -3.560 | -0.4488 | Yes | ||
| 88 | ZFAND3 | 17525 | -3.561 | -0.4335 | Yes | ||
| 89 | XBP1 | 17652 | -3.801 | -0.4240 | Yes | ||
| 90 | RANBP10 | 17744 | -3.986 | -0.4118 | Yes | ||
| 91 | PLAGL2 | 17833 | -4.192 | -0.3986 | Yes | ||
| 92 | MAP3K3 | 18061 | -4.882 | -0.3899 | Yes | ||
| 93 | SORT1 | 18071 | -4.936 | -0.3692 | Yes | ||
| 94 | KLF13 | 18072 | -4.938 | -0.3480 | Yes | ||
| 95 | PPP1R9B | 18121 | -5.135 | -0.3285 | Yes | ||
| 96 | GLCCI1 | 18181 | -5.473 | -0.3082 | Yes | ||
| 97 | BRD2 | 18187 | -5.490 | -0.2849 | Yes | ||
| 98 | LPHN1 | 18221 | -5.642 | -0.2625 | Yes | ||
| 99 | GPRASP2 | 18240 | -5.737 | -0.2388 | Yes | ||
| 100 | UHRF2 | 18295 | -6.129 | -0.2154 | Yes | ||
| 101 | UBAP1 | 18297 | -6.131 | -0.1892 | Yes | ||
| 102 | PARP6 | 18348 | -6.525 | -0.1639 | Yes | ||
| 103 | RNF44 | 18358 | -6.612 | -0.1360 | Yes | ||
| 104 | RARA | 18442 | -7.500 | -0.1083 | Yes | ||
| 105 | RGS16 | 18486 | -8.202 | -0.0754 | Yes | ||
| 106 | VAMP1 | 18525 | -9.402 | -0.0371 | Yes | ||
| 107 | PHF2 | 18536 | -9.766 | 0.0043 | Yes |
