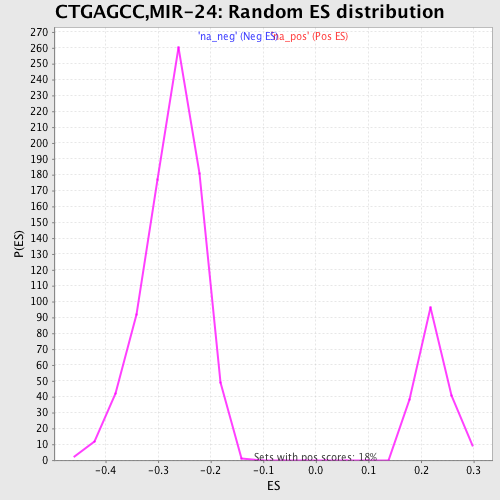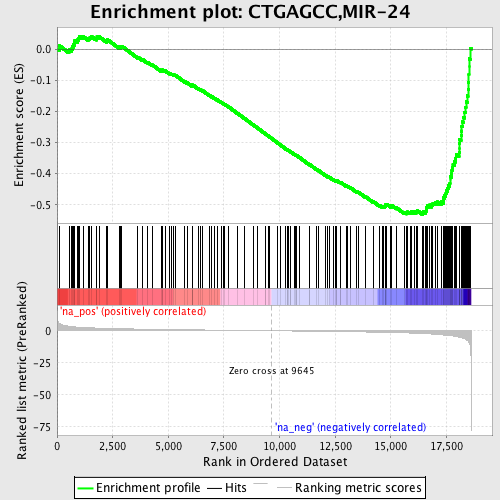
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CTGAGCC,MIR-24 |
| Enrichment Score (ES) | -0.53102094 |
| Normalized Enrichment Score (NES) | -1.9428462 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0951071 |
| FWER p-Value | 0.218 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ABCB9 | 96 | 5.709 | 0.0113 | No | ||
| 2 | AARS | 536 | 3.562 | -0.0023 | No | ||
| 3 | B3GNT6 | 662 | 3.254 | 0.0003 | No | ||
| 4 | GPX3 | 693 | 3.190 | 0.0079 | No | ||
| 5 | DOCK3 | 737 | 3.103 | 0.0145 | No | ||
| 6 | RNF138 | 779 | 3.044 | 0.0211 | No | ||
| 7 | CCT3 | 798 | 3.017 | 0.0288 | No | ||
| 8 | NEK4 | 933 | 2.820 | 0.0297 | No | ||
| 9 | XPO4 | 974 | 2.775 | 0.0355 | No | ||
| 10 | PPM1G | 1009 | 2.737 | 0.0415 | No | ||
| 11 | FBXL19 | 1172 | 2.579 | 0.0402 | No | ||
| 12 | MATR3 | 1394 | 2.367 | 0.0351 | No | ||
| 13 | MAFG | 1476 | 2.301 | 0.0373 | No | ||
| 14 | TCF2 | 1536 | 2.268 | 0.0406 | No | ||
| 15 | LENG4 | 1753 | 2.126 | 0.0351 | No | ||
| 16 | TOP1 | 1791 | 2.107 | 0.0391 | No | ||
| 17 | FGFR3 | 1889 | 2.045 | 0.0398 | No | ||
| 18 | SEMA6B | 2237 | 1.866 | 0.0264 | No | ||
| 19 | NDST1 | 2250 | 1.860 | 0.0311 | No | ||
| 20 | PIGS | 2786 | 1.618 | 0.0068 | No | ||
| 21 | TAF5 | 2867 | 1.588 | 0.0070 | No | ||
| 22 | EDA | 2908 | 1.574 | 0.0094 | No | ||
| 23 | HAP1 | 3625 | 1.327 | -0.0256 | No | ||
| 24 | EXOC5 | 3819 | 1.271 | -0.0324 | No | ||
| 25 | GRIA3 | 4054 | 1.199 | -0.0416 | No | ||
| 26 | TRIM33 | 4287 | 1.137 | -0.0509 | No | ||
| 27 | KPNA3 | 4672 | 1.044 | -0.0687 | No | ||
| 28 | PURA | 4742 | 1.026 | -0.0695 | No | ||
| 29 | PRSS8 | 4749 | 1.025 | -0.0668 | No | ||
| 30 | TNFRSF19 | 4857 | 1.000 | -0.0698 | No | ||
| 31 | PTPN9 | 5052 | 0.958 | -0.0775 | No | ||
| 32 | GPC4 | 5150 | 0.931 | -0.0801 | No | ||
| 33 | RNF11 | 5236 | 0.908 | -0.0821 | No | ||
| 34 | PDGFRA | 5317 | 0.887 | -0.0838 | No | ||
| 35 | RARG | 5744 | 0.786 | -0.1046 | No | ||
| 36 | STC2 | 5843 | 0.766 | -0.1077 | No | ||
| 37 | BSN | 6073 | 0.715 | -0.1181 | No | ||
| 38 | NEUROD1 | 6074 | 0.714 | -0.1160 | No | ||
| 39 | RNF150 | 6095 | 0.711 | -0.1151 | No | ||
| 40 | PAK4 | 6365 | 0.653 | -0.1278 | No | ||
| 41 | CDV3 | 6442 | 0.638 | -0.1300 | No | ||
| 42 | BHLHB5 | 6545 | 0.620 | -0.1338 | No | ||
| 43 | EPHA8 | 6840 | 0.560 | -0.1481 | No | ||
| 44 | PTGER4 | 6953 | 0.540 | -0.1526 | No | ||
| 45 | AMMECR1 | 7094 | 0.511 | -0.1587 | No | ||
| 46 | NEK9 | 7189 | 0.490 | -0.1624 | No | ||
| 47 | MMP16 | 7376 | 0.449 | -0.1712 | No | ||
| 48 | LYPLA2 | 7471 | 0.430 | -0.1750 | No | ||
| 49 | NELL2 | 7538 | 0.417 | -0.1774 | No | ||
| 50 | JUB | 7700 | 0.388 | -0.1850 | No | ||
| 51 | RAP2C | 8117 | 0.300 | -0.2067 | No | ||
| 52 | ACVR1B | 8440 | 0.237 | -0.2234 | No | ||
| 53 | INSIG1 | 8825 | 0.159 | -0.2438 | No | ||
| 54 | FST | 8994 | 0.125 | -0.2525 | No | ||
| 55 | SCML2 | 9353 | 0.059 | -0.2717 | No | ||
| 56 | TMEM9 | 9381 | 0.052 | -0.2731 | No | ||
| 57 | RASGRP4 | 9480 | 0.031 | -0.2783 | No | ||
| 58 | RAP1B | 9539 | 0.018 | -0.2814 | No | ||
| 59 | LRFN2 | 9896 | -0.049 | -0.3005 | No | ||
| 60 | DCBLD2 | 10033 | -0.076 | -0.3077 | No | ||
| 61 | WDR23 | 10284 | -0.126 | -0.3208 | No | ||
| 62 | HIP1R | 10347 | -0.137 | -0.3238 | No | ||
| 63 | IGFBP5 | 10363 | -0.140 | -0.3242 | No | ||
| 64 | SFXN5 | 10400 | -0.148 | -0.3257 | No | ||
| 65 | PER1 | 10468 | -0.161 | -0.3289 | No | ||
| 66 | RASA1 | 10493 | -0.165 | -0.3297 | No | ||
| 67 | ARID5B | 10661 | -0.206 | -0.3382 | No | ||
| 68 | SMCR8 | 10733 | -0.221 | -0.3414 | No | ||
| 69 | KLHL1 | 10752 | -0.227 | -0.3417 | No | ||
| 70 | DLGAP4 | 10776 | -0.232 | -0.3423 | No | ||
| 71 | COL11A2 | 10888 | -0.255 | -0.3476 | No | ||
| 72 | AMOTL2 | 11347 | -0.350 | -0.3714 | No | ||
| 73 | YBX2 | 11348 | -0.350 | -0.3704 | No | ||
| 74 | MAP3K7IP2 | 11673 | -0.421 | -0.3867 | No | ||
| 75 | APOA5 | 11764 | -0.444 | -0.3903 | No | ||
| 76 | SLC19A2 | 12072 | -0.506 | -0.4055 | No | ||
| 77 | EDG1 | 12175 | -0.527 | -0.4095 | No | ||
| 78 | RAP1A | 12241 | -0.541 | -0.4114 | No | ||
| 79 | PTPRD | 12436 | -0.588 | -0.4202 | No | ||
| 80 | TRPM6 | 12526 | -0.612 | -0.4233 | No | ||
| 81 | APBA1 | 12576 | -0.622 | -0.4242 | No | ||
| 82 | CLCN6 | 12577 | -0.623 | -0.4224 | No | ||
| 83 | CDX2 | 12719 | -0.657 | -0.4281 | No | ||
| 84 | MAPK7 | 12748 | -0.663 | -0.4277 | No | ||
| 85 | CALCR | 12994 | -0.717 | -0.4389 | No | ||
| 86 | WDTC1 | 13061 | -0.735 | -0.4404 | No | ||
| 87 | DNAJB12 | 13190 | -0.772 | -0.4451 | No | ||
| 88 | SUV420H2 | 13473 | -0.842 | -0.4579 | No | ||
| 89 | PER2 | 13542 | -0.859 | -0.4591 | No | ||
| 90 | ADCY9 | 13865 | -0.947 | -0.4739 | No | ||
| 91 | ACBD4 | 14222 | -1.052 | -0.4901 | No | ||
| 92 | DLC1 | 14487 | -1.132 | -0.5011 | No | ||
| 93 | PCDH10 | 14616 | -1.170 | -0.5047 | No | ||
| 94 | CACNB1 | 14674 | -1.190 | -0.5044 | No | ||
| 95 | PTPRF | 14738 | -1.210 | -0.5043 | No | ||
| 96 | CMTM4 | 14763 | -1.221 | -0.5021 | No | ||
| 97 | CSK | 14774 | -1.225 | -0.4991 | No | ||
| 98 | FURIN | 14825 | -1.246 | -0.4982 | No | ||
| 99 | MARK4 | 15005 | -1.319 | -0.5041 | No | ||
| 100 | ADAM19 | 15034 | -1.333 | -0.5018 | No | ||
| 101 | ATP6V0A2 | 15250 | -1.421 | -0.5093 | No | ||
| 102 | MNT | 15615 | -1.616 | -0.5244 | No | ||
| 103 | PHC1 | 15710 | -1.669 | -0.5247 | No | ||
| 104 | SP1 | 15765 | -1.699 | -0.5227 | No | ||
| 105 | SYP | 15885 | -1.763 | -0.5241 | No | ||
| 106 | INPP5B | 15936 | -1.797 | -0.5216 | No | ||
| 107 | KCNK2 | 16057 | -1.880 | -0.5227 | No | ||
| 108 | H2AFX | 16133 | -1.940 | -0.5211 | No | ||
| 109 | PRKRIP1 | 16201 | -1.984 | -0.5190 | No | ||
| 110 | CNTFR | 16423 | -2.159 | -0.5248 | Yes | ||
| 111 | DHX30 | 16476 | -2.199 | -0.5213 | Yes | ||
| 112 | PLOD2 | 16579 | -2.284 | -0.5202 | Yes | ||
| 113 | HIC2 | 16591 | -2.293 | -0.5142 | Yes | ||
| 114 | PLK3 | 16606 | -2.308 | -0.5083 | Yes | ||
| 115 | SBK1 | 16633 | -2.330 | -0.5030 | Yes | ||
| 116 | RNF2 | 16716 | -2.411 | -0.5005 | Yes | ||
| 117 | MOCS1 | 16842 | -2.550 | -0.4999 | Yes | ||
| 118 | POGZ | 16891 | -2.601 | -0.4950 | Yes | ||
| 119 | OGT | 16987 | -2.708 | -0.4924 | Yes | ||
| 120 | BCL2L11 | 17107 | -2.863 | -0.4906 | Yes | ||
| 121 | ING5 | 17264 | -3.095 | -0.4901 | Yes | ||
| 122 | GBA2 | 17361 | -3.245 | -0.4859 | Yes | ||
| 123 | RAB5C | 17378 | -3.276 | -0.4774 | Yes | ||
| 124 | MMP14 | 17421 | -3.354 | -0.4700 | Yes | ||
| 125 | PIM2 | 17476 | -3.462 | -0.4629 | Yes | ||
| 126 | SESN1 | 17514 | -3.538 | -0.4547 | Yes | ||
| 127 | ZXDA | 17558 | -3.609 | -0.4467 | Yes | ||
| 128 | VAV1 | 17585 | -3.659 | -0.4375 | Yes | ||
| 129 | SP6 | 17653 | -3.802 | -0.4302 | Yes | ||
| 130 | CLK2 | 17661 | -3.812 | -0.4196 | Yes | ||
| 131 | ALS2CR2 | 17662 | -3.813 | -0.4086 | Yes | ||
| 132 | MIDN | 17727 | -3.951 | -0.4007 | Yes | ||
| 133 | RANBP10 | 17744 | -3.986 | -0.3900 | Yes | ||
| 134 | RHBDL1 | 17758 | -4.028 | -0.3791 | Yes | ||
| 135 | MAGI1 | 17793 | -4.080 | -0.3692 | Yes | ||
| 136 | MEN1 | 17871 | -4.297 | -0.3610 | Yes | ||
| 137 | BCL2L2 | 17906 | -4.415 | -0.3501 | Yes | ||
| 138 | RGL2 | 17929 | -4.481 | -0.3384 | Yes | ||
| 139 | TOB2 | 18066 | -4.918 | -0.3316 | Yes | ||
| 140 | RASSF2 | 18078 | -4.955 | -0.3179 | Yes | ||
| 141 | PLCL2 | 18081 | -4.970 | -0.3037 | Yes | ||
| 142 | PIM1 | 18097 | -5.045 | -0.2900 | Yes | ||
| 143 | PSAP | 18157 | -5.347 | -0.2778 | Yes | ||
| 144 | RAB5B | 18176 | -5.449 | -0.2630 | Yes | ||
| 145 | PIP5K1B | 18191 | -5.499 | -0.2479 | Yes | ||
| 146 | SEMA4A | 18231 | -5.683 | -0.2337 | Yes | ||
| 147 | MXI1 | 18255 | -5.834 | -0.2181 | Yes | ||
| 148 | SSH2 | 18321 | -6.266 | -0.2036 | Yes | ||
| 149 | PARP6 | 18348 | -6.525 | -0.1862 | Yes | ||
| 150 | KIF21B | 18381 | -6.788 | -0.1683 | Yes | ||
| 151 | TRPC4AP | 18436 | -7.441 | -0.1498 | Yes | ||
| 152 | NEDD9 | 18470 | -7.962 | -0.1286 | Yes | ||
| 153 | ARHGEF5 | 18487 | -8.231 | -0.1058 | Yes | ||
| 154 | CDKN1B | 18510 | -8.697 | -0.0819 | Yes | ||
| 155 | DUSP16 | 18514 | -8.989 | -0.0562 | Yes | ||
| 156 | SLC12A6 | 18516 | -9.060 | -0.0301 | Yes | ||
| 157 | RAB6IP1 | 18567 | -12.306 | 0.0027 | Yes |