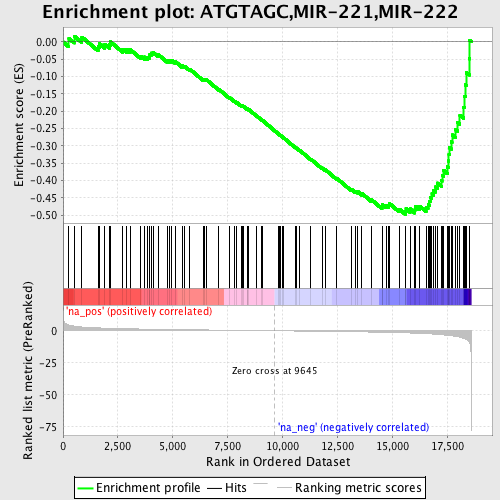
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATGTAGC,MIR-221,MIR-222 |
| Enrichment Score (ES) | -0.49760398 |
| Normalized Enrichment Score (NES) | -1.7407671 |
| Nominal p-value | 0.0025740026 |
| FDR q-value | 0.10245224 |
| FWER p-Value | 0.759 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PCDHA7 | 265 | 4.437 | 0.0100 | No | ||
| 2 | TIMP3 | 507 | 3.621 | 0.0168 | No | ||
| 3 | ANGPTL2 | 853 | 2.927 | 0.0143 | No | ||
| 4 | DCUN1D1 | 1598 | 2.222 | -0.0137 | No | ||
| 5 | CDKN1C | 1662 | 2.183 | -0.0052 | No | ||
| 6 | BMF | 1901 | 2.039 | -0.0068 | No | ||
| 7 | FNDC3A | 2115 | 1.929 | -0.0078 | No | ||
| 8 | ACACA | 2154 | 1.907 | 0.0006 | No | ||
| 9 | KHDRBS2 | 2721 | 1.639 | -0.0209 | No | ||
| 10 | UBE2J1 | 2886 | 1.583 | -0.0211 | No | ||
| 11 | PPP3R1 | 3054 | 1.521 | -0.0218 | No | ||
| 12 | TRPS1 | 3544 | 1.351 | -0.0408 | No | ||
| 13 | HMGCLL1 | 3722 | 1.299 | -0.0432 | No | ||
| 14 | IRX5 | 3865 | 1.253 | -0.0440 | No | ||
| 15 | QKI | 3943 | 1.229 | -0.0415 | No | ||
| 16 | NRK | 3954 | 1.226 | -0.0353 | No | ||
| 17 | TCF12 | 4014 | 1.210 | -0.0318 | No | ||
| 18 | LIFR | 4114 | 1.183 | -0.0307 | No | ||
| 19 | DMRT3 | 4332 | 1.128 | -0.0362 | No | ||
| 20 | PURA | 4742 | 1.026 | -0.0527 | No | ||
| 21 | RALA | 4847 | 1.002 | -0.0528 | No | ||
| 22 | ZADH2 | 4948 | 0.981 | -0.0528 | No | ||
| 23 | MAPK10 | 5114 | 0.942 | -0.0566 | No | ||
| 24 | PAIP1 | 5422 | 0.860 | -0.0684 | No | ||
| 25 | PLEKHC1 | 5528 | 0.837 | -0.0695 | No | ||
| 26 | PPARGC1A | 5777 | 0.779 | -0.0786 | No | ||
| 27 | PCDHA3 | 6380 | 0.649 | -0.1076 | No | ||
| 28 | CDV3 | 6442 | 0.638 | -0.1074 | No | ||
| 29 | NAP1L5 | 6538 | 0.621 | -0.1091 | No | ||
| 30 | AMMECR1 | 7094 | 0.511 | -0.1363 | No | ||
| 31 | PDCD10 | 7586 | 0.409 | -0.1605 | No | ||
| 32 | NTF3 | 7805 | 0.367 | -0.1703 | No | ||
| 33 | EIF5A2 | 7922 | 0.341 | -0.1747 | No | ||
| 34 | HOXC10 | 8123 | 0.298 | -0.1839 | No | ||
| 35 | SNCB | 8154 | 0.293 | -0.1839 | No | ||
| 36 | RAB1A | 8219 | 0.278 | -0.1858 | No | ||
| 37 | SHANK2 | 8387 | 0.248 | -0.1935 | No | ||
| 38 | PCDHA10 | 8392 | 0.248 | -0.1923 | No | ||
| 39 | CPNE8 | 8471 | 0.230 | -0.1953 | No | ||
| 40 | INSIG1 | 8825 | 0.159 | -0.2135 | No | ||
| 41 | ZFPM2 | 9038 | 0.118 | -0.2243 | No | ||
| 42 | MAP3K10 | 9073 | 0.112 | -0.2255 | No | ||
| 43 | RSBN1L | 9798 | -0.028 | -0.2644 | No | ||
| 44 | PCDHA5 | 9859 | -0.041 | -0.2674 | No | ||
| 45 | LRFN2 | 9896 | -0.049 | -0.2691 | No | ||
| 46 | ARF4 | 9979 | -0.066 | -0.2732 | No | ||
| 47 | YWHAG | 10062 | -0.083 | -0.2772 | No | ||
| 48 | PAIP2 | 10595 | -0.189 | -0.3049 | No | ||
| 49 | RIMS3 | 10631 | -0.199 | -0.3056 | No | ||
| 50 | PCDHA11 | 10782 | -0.233 | -0.3125 | No | ||
| 51 | PCDHA12 | 11294 | -0.339 | -0.3382 | No | ||
| 52 | FOS | 11810 | -0.455 | -0.3635 | No | ||
| 53 | HECTD2 | 11949 | -0.479 | -0.3684 | No | ||
| 54 | CTCF | 12473 | -0.599 | -0.3933 | No | ||
| 55 | DACH1 | 13144 | -0.760 | -0.4253 | No | ||
| 56 | ADAM11 | 13322 | -0.805 | -0.4305 | No | ||
| 57 | CBFB | 13422 | -0.828 | -0.4313 | No | ||
| 58 | FMR1 | 13617 | -0.878 | -0.4369 | No | ||
| 59 | EIF3S1 | 14058 | -1.000 | -0.4552 | No | ||
| 60 | ESR1 | 14537 | -1.146 | -0.4747 | No | ||
| 61 | VAPB | 14538 | -1.147 | -0.4685 | No | ||
| 62 | ATP1A1 | 14716 | -1.202 | -0.4714 | No | ||
| 63 | GARNL1 | 14827 | -1.247 | -0.4705 | No | ||
| 64 | PAK1 | 14871 | -1.265 | -0.4659 | No | ||
| 65 | SLC25A37 | 15314 | -1.450 | -0.4818 | No | ||
| 66 | SEMA6D | 15607 | -1.612 | -0.4888 | Yes | ||
| 67 | PCDHA6 | 15627 | -1.622 | -0.4809 | Yes | ||
| 68 | AXIN2 | 15813 | -1.724 | -0.4814 | Yes | ||
| 69 | HRB | 16036 | -1.869 | -0.4832 | Yes | ||
| 70 | NLK | 16066 | -1.886 | -0.4744 | Yes | ||
| 71 | MEIS1 | 16251 | -2.021 | -0.4733 | Yes | ||
| 72 | ZNF385 | 16544 | -2.248 | -0.4767 | Yes | ||
| 73 | SBK1 | 16633 | -2.330 | -0.4687 | Yes | ||
| 74 | PCDHA1 | 16709 | -2.402 | -0.4596 | Yes | ||
| 75 | VGLL4 | 16731 | -2.435 | -0.4474 | Yes | ||
| 76 | ETS1 | 16812 | -2.516 | -0.4379 | Yes | ||
| 77 | POGZ | 16891 | -2.601 | -0.4279 | Yes | ||
| 78 | OGT | 16987 | -2.708 | -0.4181 | Yes | ||
| 79 | DNAJC14 | 17062 | -2.805 | -0.4068 | Yes | ||
| 80 | ETS2 | 17249 | -3.075 | -0.4000 | Yes | ||
| 81 | ARNT | 17271 | -3.107 | -0.3841 | Yes | ||
| 82 | IRF2 | 17342 | -3.221 | -0.3702 | Yes | ||
| 83 | ASB7 | 17511 | -3.528 | -0.3599 | Yes | ||
| 84 | PPP6C | 17569 | -3.635 | -0.3431 | Yes | ||
| 85 | ARID1A | 17584 | -3.658 | -0.3238 | Yes | ||
| 86 | MYO10 | 17611 | -3.707 | -0.3049 | Yes | ||
| 87 | CD4 | 17691 | -3.887 | -0.2878 | Yes | ||
| 88 | MIDN | 17727 | -3.951 | -0.2680 | Yes | ||
| 89 | MYLIP | 17879 | -4.319 | -0.2525 | Yes | ||
| 90 | ATXN1 | 17981 | -4.640 | -0.2325 | Yes | ||
| 91 | HIPK1 | 18085 | -4.982 | -0.2108 | Yes | ||
| 92 | WDR40A | 18258 | -5.846 | -0.1880 | Yes | ||
| 93 | DYRK1A | 18303 | -6.167 | -0.1566 | Yes | ||
| 94 | TOX | 18338 | -6.406 | -0.1233 | Yes | ||
| 95 | MESDC1 | 18377 | -6.745 | -0.0884 | Yes | ||
| 96 | CDKN1B | 18510 | -8.697 | -0.0479 | Yes | ||
| 97 | PHF2 | 18536 | -9.766 | 0.0043 | Yes |
