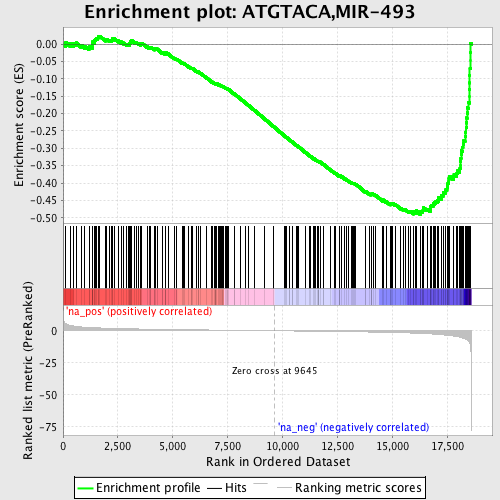
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATGTACA,MIR-493 |
| Enrichment Score (ES) | -0.49064893 |
| Normalized Enrichment Score (NES) | -1.8717943 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08787663 |
| FWER p-Value | 0.368 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SNRP70 | 124 | 5.420 | 0.0048 | No | ||
| 2 | OTUB1 | 335 | 4.183 | 0.0023 | No | ||
| 3 | CARM1 | 488 | 3.699 | 0.0019 | No | ||
| 4 | PPP2R5D | 589 | 3.429 | 0.0038 | No | ||
| 5 | PBX3 | 833 | 2.954 | -0.0031 | No | ||
| 6 | SPIRE2 | 994 | 2.752 | -0.0060 | No | ||
| 7 | ANKRD17 | 1182 | 2.571 | -0.0107 | No | ||
| 8 | ETF1 | 1215 | 2.540 | -0.0070 | No | ||
| 9 | RALGPS1 | 1329 | 2.419 | -0.0080 | No | ||
| 10 | SLC25A26 | 1334 | 2.414 | -0.0031 | No | ||
| 11 | CHKB | 1336 | 2.414 | 0.0020 | No | ||
| 12 | DDX3X | 1349 | 2.406 | 0.0065 | No | ||
| 13 | EPHA7 | 1427 | 2.337 | 0.0073 | No | ||
| 14 | NMT1 | 1440 | 2.327 | 0.0116 | No | ||
| 15 | ATAD2 | 1479 | 2.298 | 0.0144 | No | ||
| 16 | DHX36 | 1524 | 2.275 | 0.0169 | No | ||
| 17 | GABRA1 | 1590 | 2.227 | 0.0181 | No | ||
| 18 | DCUN1D1 | 1598 | 2.222 | 0.0224 | No | ||
| 19 | ARFGEF1 | 1670 | 2.177 | 0.0232 | No | ||
| 20 | BRD7 | 1934 | 2.015 | 0.0132 | No | ||
| 21 | CDH2 | 1985 | 1.994 | 0.0147 | No | ||
| 22 | ZMYND19 | 2132 | 1.924 | 0.0109 | No | ||
| 23 | PMP22 | 2212 | 1.878 | 0.0106 | No | ||
| 24 | DAB2IP | 2234 | 1.869 | 0.0134 | No | ||
| 25 | NDST1 | 2250 | 1.860 | 0.0166 | No | ||
| 26 | TTC7B | 2341 | 1.817 | 0.0155 | No | ||
| 27 | SEMA4C | 2519 | 1.731 | 0.0096 | No | ||
| 28 | SORCS3 | 2642 | 1.674 | 0.0065 | No | ||
| 29 | ATP2A2 | 2760 | 1.626 | 0.0036 | No | ||
| 30 | SVIL | 2893 | 1.580 | -0.0002 | No | ||
| 31 | RBBP6 | 2963 | 1.552 | -0.0006 | No | ||
| 32 | RPS6KA5 | 3014 | 1.536 | -0.0001 | No | ||
| 33 | CDH11 | 3032 | 1.530 | 0.0023 | No | ||
| 34 | EIF2C1 | 3056 | 1.521 | 0.0043 | No | ||
| 35 | AK5 | 3062 | 1.519 | 0.0072 | No | ||
| 36 | E2F6 | 3105 | 1.504 | 0.0081 | No | ||
| 37 | SFRS3 | 3116 | 1.498 | 0.0108 | No | ||
| 38 | GABRG2 | 3273 | 1.442 | 0.0054 | No | ||
| 39 | RSBN1 | 3327 | 1.421 | 0.0055 | No | ||
| 40 | CDC14A | 3452 | 1.380 | 0.0017 | No | ||
| 41 | GAD1 | 3539 | 1.351 | -0.0001 | No | ||
| 42 | TRPS1 | 3544 | 1.351 | 0.0026 | No | ||
| 43 | ZIC1 | 3587 | 1.338 | 0.0031 | No | ||
| 44 | CUGBP2 | 3835 | 1.266 | -0.0076 | No | ||
| 45 | MBNL2 | 3950 | 1.227 | -0.0112 | No | ||
| 46 | TTN | 3978 | 1.220 | -0.0100 | No | ||
| 47 | WNT5A | 3996 | 1.215 | -0.0084 | No | ||
| 48 | WDFY3 | 4153 | 1.173 | -0.0144 | No | ||
| 49 | UBE3A | 4181 | 1.163 | -0.0134 | No | ||
| 50 | PIAS1 | 4218 | 1.153 | -0.0129 | No | ||
| 51 | SRPX | 4281 | 1.138 | -0.0138 | No | ||
| 52 | ENAH | 4516 | 1.081 | -0.0242 | No | ||
| 53 | TMEM16D | 4551 | 1.073 | -0.0238 | No | ||
| 54 | AR | 4664 | 1.045 | -0.0276 | No | ||
| 55 | SH2D3C | 4683 | 1.041 | -0.0264 | No | ||
| 56 | SOX21 | 4685 | 1.041 | -0.0242 | No | ||
| 57 | MARK1 | 4790 | 1.017 | -0.0277 | No | ||
| 58 | MTPN | 5067 | 0.955 | -0.0407 | No | ||
| 59 | GPC4 | 5150 | 0.931 | -0.0432 | No | ||
| 60 | CCNT2 | 5179 | 0.924 | -0.0427 | No | ||
| 61 | PAIP1 | 5422 | 0.860 | -0.0541 | No | ||
| 62 | CLPX | 5479 | 0.848 | -0.0553 | No | ||
| 63 | MLLT10 | 5533 | 0.837 | -0.0564 | No | ||
| 64 | ZIC2 | 5723 | 0.792 | -0.0650 | No | ||
| 65 | PIK3R1 | 5830 | 0.769 | -0.0691 | No | ||
| 66 | ATP5A1 | 5869 | 0.760 | -0.0696 | No | ||
| 67 | SALL1 | 5900 | 0.755 | -0.0696 | No | ||
| 68 | NLGN3 | 6092 | 0.712 | -0.0784 | No | ||
| 69 | VPS13D | 6150 | 0.701 | -0.0801 | No | ||
| 70 | KPNA4 | 6163 | 0.699 | -0.0792 | No | ||
| 71 | EYA1 | 6264 | 0.675 | -0.0832 | No | ||
| 72 | BHLHB5 | 6545 | 0.620 | -0.0971 | No | ||
| 73 | UBE2G1 | 6782 | 0.574 | -0.1087 | No | ||
| 74 | SPRYD3 | 6828 | 0.563 | -0.1100 | No | ||
| 75 | NXPH1 | 6903 | 0.547 | -0.1128 | No | ||
| 76 | PSD2 | 6948 | 0.541 | -0.1141 | No | ||
| 77 | PCDH18 | 6950 | 0.541 | -0.1130 | No | ||
| 78 | UNC50 | 6957 | 0.539 | -0.1121 | No | ||
| 79 | ESRRG | 7000 | 0.529 | -0.1133 | No | ||
| 80 | ZFX | 7101 | 0.510 | -0.1177 | No | ||
| 81 | MAST1 | 7115 | 0.508 | -0.1173 | No | ||
| 82 | HEMGN | 7171 | 0.494 | -0.1192 | No | ||
| 83 | FHOD3 | 7210 | 0.485 | -0.1203 | No | ||
| 84 | SNAI1 | 7244 | 0.476 | -0.1210 | No | ||
| 85 | HIVEP2 | 7254 | 0.475 | -0.1205 | No | ||
| 86 | ANTXR1 | 7296 | 0.465 | -0.1217 | No | ||
| 87 | FZD4 | 7401 | 0.445 | -0.1265 | No | ||
| 88 | ADAM10 | 7445 | 0.435 | -0.1279 | No | ||
| 89 | HMGCS1 | 7478 | 0.429 | -0.1287 | No | ||
| 90 | EFNA3 | 7488 | 0.427 | -0.1283 | No | ||
| 91 | CDC73 | 7555 | 0.415 | -0.1310 | No | ||
| 92 | SDC2 | 7804 | 0.367 | -0.1437 | No | ||
| 93 | GABRA5 | 7806 | 0.367 | -0.1429 | No | ||
| 94 | BHLHB3 | 8100 | 0.303 | -0.1582 | No | ||
| 95 | TRAM2 | 8290 | 0.263 | -0.1679 | No | ||
| 96 | CPNE8 | 8471 | 0.230 | -0.1772 | No | ||
| 97 | TBC1D4 | 8710 | 0.182 | -0.1898 | No | ||
| 98 | KCNMA1 | 9197 | 0.091 | -0.2160 | No | ||
| 99 | PCGF2 | 9605 | 0.007 | -0.2381 | No | ||
| 100 | DKK2 | 10078 | -0.087 | -0.2636 | No | ||
| 101 | TBK1 | 10121 | -0.096 | -0.2657 | No | ||
| 102 | RPE | 10192 | -0.109 | -0.2692 | No | ||
| 103 | PDGFC | 10322 | -0.133 | -0.2760 | No | ||
| 104 | SIPA1L2 | 10324 | -0.134 | -0.2757 | No | ||
| 105 | CDH13 | 10432 | -0.154 | -0.2812 | No | ||
| 106 | EFTUD1 | 10459 | -0.159 | -0.2823 | No | ||
| 107 | RIMS3 | 10631 | -0.199 | -0.2912 | No | ||
| 108 | PPP3CB | 10698 | -0.213 | -0.2943 | No | ||
| 109 | SRPK2 | 10738 | -0.222 | -0.2960 | No | ||
| 110 | SS18 | 11056 | -0.290 | -0.3126 | No | ||
| 111 | MEF2C | 11230 | -0.324 | -0.3213 | No | ||
| 112 | NLGN1 | 11266 | -0.333 | -0.3225 | No | ||
| 113 | SP3 | 11425 | -0.367 | -0.3303 | No | ||
| 114 | VAMP2 | 11436 | -0.370 | -0.3300 | No | ||
| 115 | RHOT1 | 11518 | -0.388 | -0.3336 | No | ||
| 116 | TNFSF11 | 11598 | -0.404 | -0.3371 | No | ||
| 117 | KCNIP4 | 11624 | -0.410 | -0.3375 | No | ||
| 118 | LRRTM3 | 11631 | -0.411 | -0.3370 | No | ||
| 119 | CTDSPL | 11650 | -0.416 | -0.3371 | No | ||
| 120 | RBM12 | 11708 | -0.430 | -0.3393 | No | ||
| 121 | FBXO33 | 11868 | -0.466 | -0.3469 | No | ||
| 122 | TMEFF1 | 11889 | -0.470 | -0.3470 | No | ||
| 123 | PIP5K3 | 12201 | -0.533 | -0.3628 | No | ||
| 124 | UTX | 12350 | -0.567 | -0.3696 | No | ||
| 125 | UNC5A | 12428 | -0.587 | -0.3726 | No | ||
| 126 | ZBTB11 | 12586 | -0.625 | -0.3798 | No | ||
| 127 | FIGN | 12587 | -0.626 | -0.3784 | No | ||
| 128 | ITGB1 | 12596 | -0.629 | -0.3775 | No | ||
| 129 | OSBPL6 | 12710 | -0.655 | -0.3823 | No | ||
| 130 | DLL1 | 12845 | -0.683 | -0.3881 | No | ||
| 131 | NRXN3 | 12908 | -0.699 | -0.3900 | No | ||
| 132 | OVOL1 | 13021 | -0.725 | -0.3945 | No | ||
| 133 | DACH1 | 13144 | -0.760 | -0.3995 | No | ||
| 134 | CREBBP | 13192 | -0.773 | -0.4005 | No | ||
| 135 | IL1A | 13219 | -0.781 | -0.4002 | No | ||
| 136 | SPAG8 | 13266 | -0.791 | -0.4010 | No | ||
| 137 | CLASP1 | 13332 | -0.808 | -0.4028 | No | ||
| 138 | ARHGAP5 | 13771 | -0.919 | -0.4247 | No | ||
| 139 | JPH1 | 13790 | -0.924 | -0.4237 | No | ||
| 140 | TJP1 | 13981 | -0.982 | -0.4319 | No | ||
| 141 | POU4F1 | 14033 | -0.994 | -0.4326 | No | ||
| 142 | CUL5 | 14054 | -1.000 | -0.4315 | No | ||
| 143 | NUDT4 | 14076 | -1.005 | -0.4305 | No | ||
| 144 | PUM2 | 14140 | -1.025 | -0.4318 | No | ||
| 145 | JUN | 14245 | -1.057 | -0.4352 | No | ||
| 146 | SIN3A | 14571 | -1.154 | -0.4504 | No | ||
| 147 | CRSP6 | 14586 | -1.161 | -0.4487 | No | ||
| 148 | AP3S1 | 14719 | -1.203 | -0.4533 | No | ||
| 149 | BCL7A | 14905 | -1.277 | -0.4606 | No | ||
| 150 | SV2B | 14912 | -1.280 | -0.4582 | No | ||
| 151 | DNAJC13 | 14977 | -1.305 | -0.4589 | No | ||
| 152 | RYBP | 15007 | -1.320 | -0.4577 | No | ||
| 153 | PDZRN3 | 15155 | -1.381 | -0.4628 | No | ||
| 154 | IRF7 | 15384 | -1.486 | -0.4720 | No | ||
| 155 | BASP1 | 15524 | -1.571 | -0.4762 | No | ||
| 156 | GRM5 | 15589 | -1.603 | -0.4763 | No | ||
| 157 | USP21 | 15738 | -1.683 | -0.4807 | No | ||
| 158 | BCOR | 15828 | -1.731 | -0.4819 | No | ||
| 159 | COBLL1 | 15967 | -1.816 | -0.4855 | No | ||
| 160 | MAPK1 | 15982 | -1.834 | -0.4824 | No | ||
| 161 | ITSN2 | 16079 | -1.895 | -0.4836 | No | ||
| 162 | CSNK1A1 | 16088 | -1.901 | -0.4799 | No | ||
| 163 | UBE2V2 | 16286 | -2.057 | -0.4863 | Yes | ||
| 164 | XPO7 | 16297 | -2.064 | -0.4824 | Yes | ||
| 165 | CRSP7 | 16368 | -2.110 | -0.4817 | Yes | ||
| 166 | APLP2 | 16399 | -2.140 | -0.4788 | Yes | ||
| 167 | SP4 | 16413 | -2.149 | -0.4749 | Yes | ||
| 168 | IRX3 | 16431 | -2.163 | -0.4713 | Yes | ||
| 169 | HIC2 | 16591 | -2.293 | -0.4750 | Yes | ||
| 170 | VGLL4 | 16731 | -2.435 | -0.4774 | Yes | ||
| 171 | GNB1 | 16756 | -2.461 | -0.4734 | Yes | ||
| 172 | CITED2 | 16757 | -2.461 | -0.4682 | Yes | ||
| 173 | FMNL2 | 16769 | -2.475 | -0.4635 | Yes | ||
| 174 | THRAP1 | 16863 | -2.571 | -0.4631 | Yes | ||
| 175 | LRP12 | 16865 | -2.572 | -0.4577 | Yes | ||
| 176 | BAZ2A | 16929 | -2.646 | -0.4555 | Yes | ||
| 177 | SNX9 | 16988 | -2.708 | -0.4529 | Yes | ||
| 178 | HNRPA0 | 17063 | -2.805 | -0.4509 | Yes | ||
| 179 | KIF1B | 17100 | -2.851 | -0.4468 | Yes | ||
| 180 | PTPN12 | 17120 | -2.878 | -0.4417 | Yes | ||
| 181 | EIF4ENIF1 | 17236 | -3.059 | -0.4415 | Yes | ||
| 182 | UBE2Q1 | 17250 | -3.076 | -0.4356 | Yes | ||
| 183 | KLF3 | 17317 | -3.169 | -0.4324 | Yes | ||
| 184 | IRF2 | 17342 | -3.221 | -0.4269 | Yes | ||
| 185 | NCOA1 | 17413 | -3.345 | -0.4236 | Yes | ||
| 186 | HS3ST3B1 | 17447 | -3.407 | -0.4181 | Yes | ||
| 187 | FOXO1A | 17497 | -3.500 | -0.4133 | Yes | ||
| 188 | NAB1 | 17518 | -3.549 | -0.4069 | Yes | ||
| 189 | PPP2R5C | 17536 | -3.576 | -0.4002 | Yes | ||
| 190 | PPP1R10 | 17579 | -3.648 | -0.3947 | Yes | ||
| 191 | ARID1A | 17584 | -3.658 | -0.3871 | Yes | ||
| 192 | SERTAD2 | 17612 | -3.708 | -0.3807 | Yes | ||
| 193 | DOCK9 | 17791 | -4.078 | -0.3817 | Yes | ||
| 194 | AFF3 | 17814 | -4.149 | -0.3740 | Yes | ||
| 195 | RNF38 | 17945 | -4.531 | -0.3715 | Yes | ||
| 196 | PHF12 | 17991 | -4.670 | -0.3640 | Yes | ||
| 197 | HIPK1 | 18085 | -4.982 | -0.3584 | Yes | ||
| 198 | FBS1 | 18099 | -5.050 | -0.3484 | Yes | ||
| 199 | SPRED2 | 18104 | -5.067 | -0.3378 | Yes | ||
| 200 | GTF2IRD1 | 18127 | -5.174 | -0.3280 | Yes | ||
| 201 | JARID1B | 18138 | -5.235 | -0.3174 | Yes | ||
| 202 | FGF13 | 18155 | -5.337 | -0.3069 | Yes | ||
| 203 | LRIG1 | 18212 | -5.590 | -0.2980 | Yes | ||
| 204 | MCL1 | 18226 | -5.653 | -0.2867 | Yes | ||
| 205 | PDPK1 | 18265 | -5.919 | -0.2761 | Yes | ||
| 206 | SSH2 | 18321 | -6.266 | -0.2658 | Yes | ||
| 207 | AXIN1 | 18349 | -6.535 | -0.2533 | Yes | ||
| 208 | MADD | 18362 | -6.643 | -0.2398 | Yes | ||
| 209 | PDE4D | 18382 | -6.789 | -0.2264 | Yes | ||
| 210 | KBTBD2 | 18398 | -6.975 | -0.2124 | Yes | ||
| 211 | PFN2 | 18413 | -7.166 | -0.1979 | Yes | ||
| 212 | ANKRD50 | 18437 | -7.443 | -0.1833 | Yes | ||
| 213 | BTG1 | 18488 | -8.254 | -0.1684 | Yes | ||
| 214 | DUSP16 | 18514 | -8.989 | -0.1507 | Yes | ||
| 215 | THRAP2 | 18517 | -9.120 | -0.1313 | Yes | ||
| 216 | BCL2 | 18530 | -9.542 | -0.1117 | Yes | ||
| 217 | PHF2 | 18536 | -9.766 | -0.0912 | Yes | ||
| 218 | SPEN | 18544 | -10.116 | -0.0700 | Yes | ||
| 219 | GAB1 | 18560 | -10.871 | -0.0477 | Yes | ||
| 220 | CAPN3 | 18562 | -11.027 | -0.0242 | Yes | ||
| 221 | TLE4 | 18570 | -12.734 | 0.0025 | Yes |
