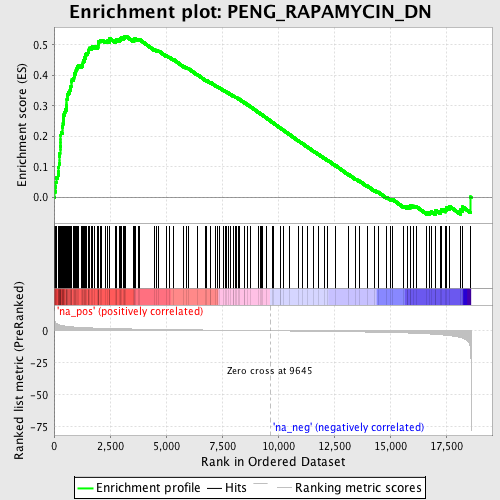
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | PENG_RAPAMYCIN_DN |
| Enrichment Score (ES) | 0.52903575 |
| Normalized Enrichment Score (NES) | 2.3867292 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SLC7A5 | 31 | 7.458 | 0.0186 | Yes | ||
| 2 | SLC29A1 | 63 | 6.281 | 0.0339 | Yes | ||
| 3 | PFKM | 79 | 6.059 | 0.0495 | Yes | ||
| 4 | GOT2 | 92 | 5.775 | 0.0646 | Yes | ||
| 5 | CYC1 | 173 | 4.902 | 0.0735 | Yes | ||
| 6 | SNRPA1 | 193 | 4.757 | 0.0854 | Yes | ||
| 7 | C1QBP | 201 | 4.728 | 0.0978 | Yes | ||
| 8 | TOMM40 | 218 | 4.634 | 0.1095 | Yes | ||
| 9 | EBNA1BP2 | 236 | 4.558 | 0.1210 | Yes | ||
| 10 | TCP1 | 237 | 4.557 | 0.1333 | Yes | ||
| 11 | GORASP2 | 253 | 4.492 | 0.1447 | Yes | ||
| 12 | ASNS | 263 | 4.441 | 0.1563 | Yes | ||
| 13 | NARS | 266 | 4.436 | 0.1682 | Yes | ||
| 14 | EIF3S9 | 292 | 4.318 | 0.1786 | Yes | ||
| 15 | PA2G4 | 296 | 4.312 | 0.1901 | Yes | ||
| 16 | FASN | 299 | 4.305 | 0.2017 | Yes | ||
| 17 | IDH3A | 307 | 4.288 | 0.2129 | Yes | ||
| 18 | NME1 | 371 | 4.010 | 0.2204 | Yes | ||
| 19 | PSMD14 | 372 | 4.009 | 0.2313 | Yes | ||
| 20 | GCN5L2 | 395 | 3.935 | 0.2407 | Yes | ||
| 21 | AATF | 406 | 3.903 | 0.2508 | Yes | ||
| 22 | MRPL12 | 419 | 3.849 | 0.2606 | Yes | ||
| 23 | NOL1 | 435 | 3.818 | 0.2701 | Yes | ||
| 24 | CDK4 | 450 | 3.776 | 0.2796 | Yes | ||
| 25 | ATF5 | 491 | 3.695 | 0.2875 | Yes | ||
| 26 | ETFA | 540 | 3.558 | 0.2945 | Yes | ||
| 27 | BCS1L | 563 | 3.501 | 0.3028 | Yes | ||
| 28 | PGK1 | 570 | 3.491 | 0.3120 | Yes | ||
| 29 | UBE2M | 571 | 3.487 | 0.3214 | Yes | ||
| 30 | SDHB | 594 | 3.417 | 0.3295 | Yes | ||
| 31 | PSMD8 | 600 | 3.404 | 0.3385 | Yes | ||
| 32 | RARS | 641 | 3.295 | 0.3452 | Yes | ||
| 33 | CCT6A | 706 | 3.163 | 0.3503 | Yes | ||
| 34 | USP10 | 727 | 3.118 | 0.3577 | Yes | ||
| 35 | SRM | 745 | 3.093 | 0.3652 | Yes | ||
| 36 | HSPA4 | 760 | 3.074 | 0.3728 | Yes | ||
| 37 | GM2A | 761 | 3.074 | 0.3811 | Yes | ||
| 38 | DDX21 | 797 | 3.018 | 0.3874 | Yes | ||
| 39 | RAE1 | 871 | 2.894 | 0.3913 | Yes | ||
| 40 | SYNCRIP | 888 | 2.876 | 0.3982 | Yes | ||
| 41 | CCT4 | 902 | 2.857 | 0.4053 | Yes | ||
| 42 | PGAM1 | 938 | 2.810 | 0.4110 | Yes | ||
| 43 | TFRC | 975 | 2.772 | 0.4166 | Yes | ||
| 44 | TARS | 1015 | 2.732 | 0.4219 | Yes | ||
| 45 | HYOU1 | 1033 | 2.719 | 0.4283 | Yes | ||
| 46 | CCNC | 1076 | 2.677 | 0.4333 | Yes | ||
| 47 | ETF1 | 1215 | 2.540 | 0.4327 | Yes | ||
| 48 | CCT2 | 1265 | 2.489 | 0.4368 | Yes | ||
| 49 | YARS | 1288 | 2.461 | 0.4423 | Yes | ||
| 50 | CALU | 1300 | 2.449 | 0.4483 | Yes | ||
| 51 | DDX3X | 1349 | 2.406 | 0.4523 | Yes | ||
| 52 | AMD1 | 1352 | 2.404 | 0.4587 | Yes | ||
| 53 | CYCS | 1401 | 2.360 | 0.4625 | Yes | ||
| 54 | SLC3A2 | 1409 | 2.354 | 0.4685 | Yes | ||
| 55 | NMT1 | 1440 | 2.327 | 0.4732 | Yes | ||
| 56 | ATP5G3 | 1512 | 2.282 | 0.4755 | Yes | ||
| 57 | CTSC | 1528 | 2.274 | 0.4809 | Yes | ||
| 58 | ADRM1 | 1556 | 2.251 | 0.4855 | Yes | ||
| 59 | WBSCR22 | 1585 | 2.229 | 0.4900 | Yes | ||
| 60 | TIMM17A | 1661 | 2.183 | 0.4919 | Yes | ||
| 61 | PSMC2 | 1712 | 2.153 | 0.4950 | Yes | ||
| 62 | AK2 | 1819 | 2.089 | 0.4949 | Yes | ||
| 63 | SCD | 1957 | 2.007 | 0.4930 | Yes | ||
| 64 | CCT5 | 1962 | 2.005 | 0.4982 | Yes | ||
| 65 | PSMB5 | 1964 | 2.005 | 0.5036 | Yes | ||
| 66 | NOL5A | 1991 | 1.991 | 0.5076 | Yes | ||
| 67 | INPP5E | 1993 | 1.990 | 0.5129 | Yes | ||
| 68 | TRIP13 | 2053 | 1.960 | 0.5150 | Yes | ||
| 69 | UBE2L3 | 2129 | 1.925 | 0.5162 | Yes | ||
| 70 | SQLE | 2292 | 1.841 | 0.5124 | Yes | ||
| 71 | HSPA5 | 2366 | 1.806 | 0.5133 | Yes | ||
| 72 | DHCR24 | 2472 | 1.755 | 0.5124 | Yes | ||
| 73 | MRPS12 | 2483 | 1.749 | 0.5166 | Yes | ||
| 74 | PSMD11 | 2485 | 1.748 | 0.5213 | Yes | ||
| 75 | PITPNB | 2748 | 1.630 | 0.5115 | Yes | ||
| 76 | ACAT2 | 2775 | 1.621 | 0.5145 | Yes | ||
| 77 | SF3A3 | 2790 | 1.616 | 0.5181 | Yes | ||
| 78 | EIF2B4 | 2921 | 1.568 | 0.5153 | Yes | ||
| 79 | CSE1L | 2961 | 1.552 | 0.5174 | Yes | ||
| 80 | RBBP6 | 2963 | 1.552 | 0.5216 | Yes | ||
| 81 | WNT10B | 2989 | 1.541 | 0.5244 | Yes | ||
| 82 | EIF4G2 | 3090 | 1.507 | 0.5231 | Yes | ||
| 83 | IDH3B | 3152 | 1.484 | 0.5238 | Yes | ||
| 84 | ARMET | 3155 | 1.483 | 0.5277 | Yes | ||
| 85 | SREBF1 | 3205 | 1.467 | 0.5290 | Yes | ||
| 86 | EIF2B2 | 3550 | 1.349 | 0.5140 | No | ||
| 87 | HSPE1 | 3571 | 1.343 | 0.5166 | No | ||
| 88 | EIF4G1 | 3577 | 1.342 | 0.5200 | No | ||
| 89 | NP | 3620 | 1.330 | 0.5213 | No | ||
| 90 | BMP1 | 3760 | 1.288 | 0.5173 | No | ||
| 91 | EMP3 | 3815 | 1.272 | 0.5178 | No | ||
| 92 | CAPZA2 | 4487 | 1.089 | 0.4844 | No | ||
| 93 | GLO1 | 4586 | 1.066 | 0.4819 | No | ||
| 94 | KPNA3 | 4672 | 1.044 | 0.4802 | No | ||
| 95 | FKBP4 | 5009 | 0.969 | 0.4646 | No | ||
| 96 | COX7B | 5144 | 0.933 | 0.4598 | No | ||
| 97 | PTBP1 | 5324 | 0.885 | 0.4525 | No | ||
| 98 | MORF4L2 | 5791 | 0.776 | 0.4294 | No | ||
| 99 | IFI30 | 5905 | 0.754 | 0.4253 | No | ||
| 100 | CTPS | 5985 | 0.735 | 0.4230 | No | ||
| 101 | TDG | 6382 | 0.649 | 0.4033 | No | ||
| 102 | GTF2A2 | 6775 | 0.575 | 0.3836 | No | ||
| 103 | PTDSS1 | 6823 | 0.564 | 0.3826 | No | ||
| 104 | IDI1 | 6988 | 0.532 | 0.3752 | No | ||
| 105 | TUBG1 | 6994 | 0.531 | 0.3763 | No | ||
| 106 | PRDX3 | 7212 | 0.484 | 0.3659 | No | ||
| 107 | NDUFA9 | 7272 | 0.470 | 0.3639 | No | ||
| 108 | MLF2 | 7378 | 0.448 | 0.3595 | No | ||
| 109 | EBP | 7554 | 0.415 | 0.3511 | No | ||
| 110 | PICALM | 7644 | 0.398 | 0.3474 | No | ||
| 111 | RTCD1 | 7702 | 0.387 | 0.3453 | No | ||
| 112 | COPB2 | 7772 | 0.374 | 0.3426 | No | ||
| 113 | ATP1B3 | 7870 | 0.351 | 0.3383 | No | ||
| 114 | AHSA1 | 8002 | 0.325 | 0.3321 | No | ||
| 115 | ICT1 | 8010 | 0.324 | 0.3326 | No | ||
| 116 | CDC25A | 8094 | 0.305 | 0.3289 | No | ||
| 117 | HCCS | 8128 | 0.297 | 0.3279 | No | ||
| 118 | RAB1A | 8219 | 0.278 | 0.3238 | No | ||
| 119 | MAPKAPK3 | 8256 | 0.270 | 0.3226 | No | ||
| 120 | PPIB | 8507 | 0.223 | 0.3096 | No | ||
| 121 | PTRF | 8520 | 0.221 | 0.3096 | No | ||
| 122 | GCDH | 8652 | 0.194 | 0.3030 | No | ||
| 123 | PSMD5 | 8760 | 0.172 | 0.2977 | No | ||
| 124 | CHUK | 9136 | 0.102 | 0.2776 | No | ||
| 125 | ATP6V0C | 9221 | 0.085 | 0.2733 | No | ||
| 126 | VCP | 9269 | 0.078 | 0.2710 | No | ||
| 127 | ARF1 | 9284 | 0.074 | 0.2704 | No | ||
| 128 | WDR18 | 9499 | 0.027 | 0.2589 | No | ||
| 129 | BTK | 9731 | -0.016 | 0.2464 | No | ||
| 130 | PPIF | 9807 | -0.029 | 0.2424 | No | ||
| 131 | ACVRL1 | 10103 | -0.091 | 0.2267 | No | ||
| 132 | BCAP31 | 10105 | -0.092 | 0.2268 | No | ||
| 133 | EEF1E1 | 10242 | -0.119 | 0.2198 | No | ||
| 134 | NUP98 | 10524 | -0.172 | 0.2050 | No | ||
| 135 | RPS26 | 10903 | -0.258 | 0.1852 | No | ||
| 136 | RNASEH1 | 11097 | -0.298 | 0.1756 | No | ||
| 137 | GSPT1 | 11321 | -0.345 | 0.1644 | No | ||
| 138 | S100A4 | 11583 | -0.401 | 0.1514 | No | ||
| 139 | BYSL | 11816 | -0.457 | 0.1400 | No | ||
| 140 | COX11 | 12078 | -0.507 | 0.1273 | No | ||
| 141 | PHB | 12202 | -0.533 | 0.1220 | No | ||
| 142 | ALDH4A1 | 12570 | -0.621 | 0.1038 | No | ||
| 143 | RABGGTB | 13147 | -0.760 | 0.0747 | No | ||
| 144 | P2RX5 | 13471 | -0.841 | 0.0594 | No | ||
| 145 | HSPA8 | 13628 | -0.880 | 0.0534 | No | ||
| 146 | TIMM44 | 13979 | -0.982 | 0.0370 | No | ||
| 147 | POLR2B | 14309 | -1.078 | 0.0221 | No | ||
| 148 | MYCBP | 14459 | -1.122 | 0.0171 | No | ||
| 149 | DAP3 | 14840 | -1.253 | -0.0001 | No | ||
| 150 | PTPN1 | 15017 | -1.323 | -0.0060 | No | ||
| 151 | NDUFS3 | 15097 | -1.356 | -0.0067 | No | ||
| 152 | CHERP | 15608 | -1.613 | -0.0299 | No | ||
| 153 | JTB | 15769 | -1.701 | -0.0340 | No | ||
| 154 | PAPOLA | 15775 | -1.703 | -0.0296 | No | ||
| 155 | NXF1 | 15889 | -1.765 | -0.0310 | No | ||
| 156 | RAB14 | 15892 | -1.768 | -0.0263 | No | ||
| 157 | SLC1A5 | 16034 | -1.867 | -0.0289 | No | ||
| 158 | XPO6 | 16156 | -1.957 | -0.0301 | No | ||
| 159 | LIMK2 | 16635 | -2.331 | -0.0497 | No | ||
| 160 | DNAJA1 | 16754 | -2.460 | -0.0494 | No | ||
| 161 | SLC39A14 | 16826 | -2.525 | -0.0464 | No | ||
| 162 | UCHL1 | 17018 | -2.743 | -0.0493 | No | ||
| 163 | NPM3 | 17025 | -2.762 | -0.0422 | No | ||
| 164 | MMP15 | 17261 | -3.092 | -0.0465 | No | ||
| 165 | RTN4 | 17290 | -3.130 | -0.0396 | No | ||
| 166 | PPP2R2A | 17459 | -3.424 | -0.0394 | No | ||
| 167 | CD47 | 17520 | -3.553 | -0.0330 | No | ||
| 168 | XBP1 | 17652 | -3.801 | -0.0298 | No | ||
| 169 | GTF3C1 | 18133 | -5.190 | -0.0417 | No | ||
| 170 | EXTL3 | 18237 | -5.720 | -0.0318 | No | ||
| 171 | PIP5K1A | 18602 | -19.271 | 0.0008 | No |
