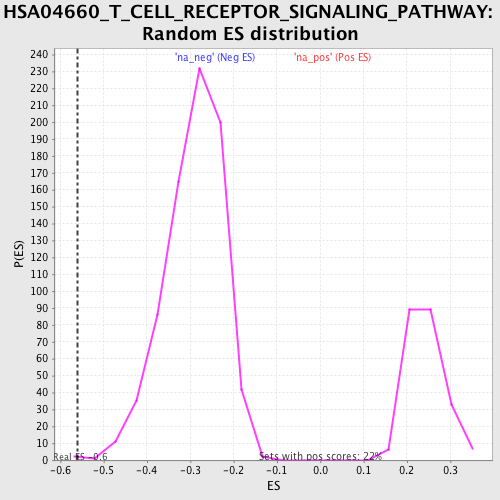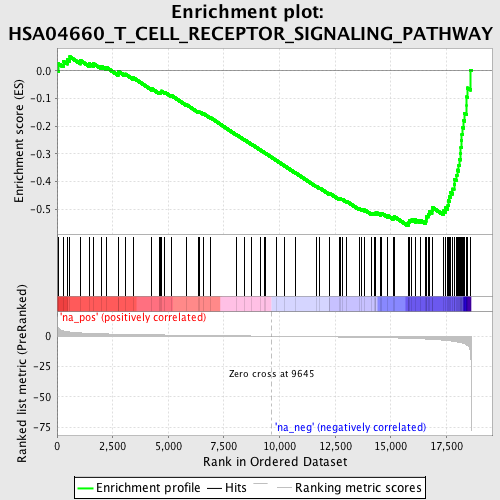
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.56038 |
| Normalized Enrichment Score (NES) | -1.919733 |
| Nominal p-value | 0.0012886598 |
| FDR q-value | 0.09566891 |
| FWER p-Value | 0.817 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL5 | 54 | 6.551 | 0.0266 | No | ||
| 2 | PIK3R3 | 276 | 4.399 | 0.0344 | No | ||
| 3 | CDK4 | 450 | 3.776 | 0.0421 | No | ||
| 4 | NFKBIE | 575 | 3.473 | 0.0510 | No | ||
| 5 | NFKBIB | 1055 | 2.697 | 0.0373 | No | ||
| 6 | PIK3R2 | 1444 | 2.324 | 0.0268 | No | ||
| 7 | GRB2 | 1642 | 2.197 | 0.0261 | No | ||
| 8 | PTPRC | 1973 | 2.001 | 0.0172 | No | ||
| 9 | NFATC2 | 2224 | 1.873 | 0.0122 | No | ||
| 10 | HRAS | 2740 | 1.631 | -0.0083 | No | ||
| 11 | CDC42 | 2751 | 1.629 | -0.0015 | No | ||
| 12 | PPP3R1 | 3054 | 1.521 | -0.0109 | No | ||
| 13 | SOS1 | 3421 | 1.389 | -0.0244 | No | ||
| 14 | MALT1 | 4259 | 1.142 | -0.0645 | No | ||
| 15 | TNF | 4598 | 1.062 | -0.0780 | No | ||
| 16 | PPP3CC | 4665 | 1.045 | -0.0768 | No | ||
| 17 | NCK1 | 4671 | 1.044 | -0.0724 | No | ||
| 18 | SOS2 | 4831 | 1.008 | -0.0764 | No | ||
| 19 | IL4 | 5143 | 0.933 | -0.0890 | No | ||
| 20 | PIK3R1 | 5830 | 0.769 | -0.1226 | No | ||
| 21 | PAK4 | 6365 | 0.653 | -0.1485 | No | ||
| 22 | PPP3R2 | 6415 | 0.645 | -0.1482 | No | ||
| 23 | NFKB1 | 6591 | 0.610 | -0.1549 | No | ||
| 24 | AKT1 | 6886 | 0.551 | -0.1683 | No | ||
| 25 | KRAS | 8061 | 0.313 | -0.2303 | No | ||
| 26 | PDCD1 | 8428 | 0.242 | -0.2489 | No | ||
| 27 | NCK2 | 8725 | 0.180 | -0.2641 | No | ||
| 28 | CHUK | 9136 | 0.102 | -0.2858 | No | ||
| 29 | CD40LG | 9328 | 0.064 | -0.2958 | No | ||
| 30 | NRAS | 9380 | 0.052 | -0.2983 | No | ||
| 31 | PIK3CA | 9878 | -0.045 | -0.3249 | No | ||
| 32 | PAK3 | 10206 | -0.112 | -0.3421 | No | ||
| 33 | PPP3CB | 10698 | -0.213 | -0.3676 | No | ||
| 34 | CBLC | 11636 | -0.413 | -0.4163 | No | ||
| 35 | FOS | 11810 | -0.455 | -0.4236 | No | ||
| 36 | NFATC4 | 12237 | -0.541 | -0.4442 | No | ||
| 37 | IKBKB | 12258 | -0.546 | -0.4428 | No | ||
| 38 | ZAP70 | 12681 | -0.648 | -0.4627 | No | ||
| 39 | CSF2 | 12718 | -0.657 | -0.4617 | No | ||
| 40 | PAK7 | 12808 | -0.676 | -0.4634 | No | ||
| 41 | IL2 | 13006 | -0.720 | -0.4708 | No | ||
| 42 | PAK2 | 13592 | -0.871 | -0.4985 | No | ||
| 43 | CD3D | 13681 | -0.894 | -0.4992 | No | ||
| 44 | PTPN6 | 13818 | -0.932 | -0.5024 | No | ||
| 45 | GRAP2 | 14115 | -1.018 | -0.5138 | No | ||
| 46 | JUN | 14245 | -1.057 | -0.5160 | No | ||
| 47 | CBLB | 14325 | -1.083 | -0.5153 | No | ||
| 48 | PLCG1 | 14331 | -1.084 | -0.5107 | No | ||
| 49 | LAT | 14531 | -1.143 | -0.5163 | No | ||
| 50 | PPP3CA | 14583 | -1.160 | -0.5139 | No | ||
| 51 | PAK1 | 14871 | -1.265 | -0.5237 | No | ||
| 52 | PIK3CG | 15103 | -1.358 | -0.5300 | No | ||
| 53 | IFNG | 15149 | -1.377 | -0.5263 | No | ||
| 54 | PIK3CB | 15782 | -1.708 | -0.5527 | Yes | ||
| 55 | PDK1 | 15809 | -1.722 | -0.5464 | Yes | ||
| 56 | IL10 | 15830 | -1.735 | -0.5396 | Yes | ||
| 57 | NFKB2 | 15949 | -1.805 | -0.5379 | Yes | ||
| 58 | MAP3K14 | 16110 | -1.919 | -0.5379 | Yes | ||
| 59 | VAV3 | 16312 | -2.074 | -0.5394 | Yes | ||
| 60 | PIK3CD | 16562 | -2.267 | -0.5426 | Yes | ||
| 61 | AKT2 | 16584 | -2.289 | -0.5335 | Yes | ||
| 62 | NFAT5 | 16614 | -2.316 | -0.5246 | Yes | ||
| 63 | VAV2 | 16677 | -2.369 | -0.5173 | Yes | ||
| 64 | CTLA4 | 16721 | -2.425 | -0.5087 | Yes | ||
| 65 | LCK | 16851 | -2.559 | -0.5042 | Yes | ||
| 66 | CD3G | 16861 | -2.568 | -0.4931 | Yes | ||
| 67 | PRKCQ | 17373 | -3.270 | -0.5060 | Yes | ||
| 68 | BCL10 | 17457 | -3.421 | -0.4951 | Yes | ||
| 69 | NFKBIA | 17567 | -3.627 | -0.4846 | Yes | ||
| 70 | VAV1 | 17585 | -3.659 | -0.4691 | Yes | ||
| 71 | MAP3K8 | 17644 | -3.789 | -0.4552 | Yes | ||
| 72 | CD4 | 17691 | -3.887 | -0.4402 | Yes | ||
| 73 | IKBKG | 17782 | -4.070 | -0.4267 | Yes | ||
| 74 | RASGRP1 | 17841 | -4.206 | -0.4109 | Yes | ||
| 75 | FYN | 17854 | -4.233 | -0.3925 | Yes | ||
| 76 | PIK3R5 | 17940 | -4.516 | -0.3768 | Yes | ||
| 77 | LCP2 | 17979 | -4.636 | -0.3580 | Yes | ||
| 78 | NFATC1 | 18056 | -4.862 | -0.3402 | Yes | ||
| 79 | NFATC3 | 18098 | -5.049 | -0.3197 | Yes | ||
| 80 | ICOS | 18141 | -5.253 | -0.2983 | Yes | ||
| 81 | CBL | 18150 | -5.305 | -0.2749 | Yes | ||
| 82 | RHOA | 18172 | -5.441 | -0.2516 | Yes | ||
| 83 | CARD11 | 18195 | -5.524 | -0.2279 | Yes | ||
| 84 | CD8B | 18236 | -5.717 | -0.2043 | Yes | ||
| 85 | CD3E | 18282 | -6.048 | -0.1796 | Yes | ||
| 86 | CD247 | 18319 | -6.255 | -0.1534 | Yes | ||
| 87 | CD28 | 18416 | -7.191 | -0.1262 | Yes | ||
| 88 | AKT3 | 18419 | -7.198 | -0.0939 | Yes | ||
| 89 | ITK | 18446 | -7.525 | -0.0615 | Yes | ||
| 90 | TEC | 18587 | -15.689 | 0.0016 | Yes |