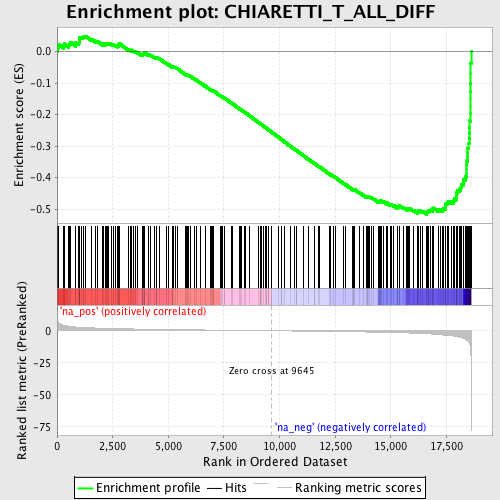
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHIARETTI_T_ALL_DIFF |
| Enrichment Score (ES) | -0.51684535 |
| Normalized Enrichment Score (NES) | -1.9731898 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.106640644 |
| FWER p-Value | 0.659 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LGALS1 | 27 | 7.814 | 0.0131 | No | ||
| 2 | CEBPB | 67 | 6.161 | 0.0225 | No | ||
| 3 | NCF4 | 302 | 4.303 | 0.0178 | No | ||
| 4 | BUB1B | 313 | 4.263 | 0.0252 | No | ||
| 5 | DDIT4 | 526 | 3.582 | 0.0203 | No | ||
| 6 | NFKBIE | 575 | 3.473 | 0.0242 | No | ||
| 7 | GCH1 | 598 | 3.411 | 0.0294 | No | ||
| 8 | GNB2L1 | 822 | 2.973 | 0.0228 | No | ||
| 9 | PBX3 | 833 | 2.954 | 0.0278 | No | ||
| 10 | USP1 | 954 | 2.794 | 0.0264 | No | ||
| 11 | CUL1 | 985 | 2.758 | 0.0300 | No | ||
| 12 | UBE2E3 | 992 | 2.754 | 0.0348 | No | ||
| 13 | PF4 | 993 | 2.752 | 0.0399 | No | ||
| 14 | IL7R | 1002 | 2.746 | 0.0446 | No | ||
| 15 | MELK | 1085 | 2.665 | 0.0451 | No | ||
| 16 | NFIL3 | 1164 | 2.587 | 0.0457 | No | ||
| 17 | STAM | 1196 | 2.558 | 0.0488 | No | ||
| 18 | MYC | 1285 | 2.465 | 0.0486 | No | ||
| 19 | GAS2 | 1552 | 2.254 | 0.0383 | No | ||
| 20 | SWAP70 | 1718 | 2.149 | 0.0334 | No | ||
| 21 | AQP3 | 1824 | 2.084 | 0.0315 | No | ||
| 22 | HIST1H4C | 2054 | 1.959 | 0.0227 | No | ||
| 23 | HNRPDL | 2082 | 1.943 | 0.0249 | No | ||
| 24 | LYN | 2171 | 1.899 | 0.0237 | No | ||
| 25 | PTPRE | 2228 | 1.871 | 0.0241 | No | ||
| 26 | ACSL1 | 2261 | 1.857 | 0.0258 | No | ||
| 27 | ITPR3 | 2321 | 1.829 | 0.0260 | No | ||
| 28 | MX1 | 2424 | 1.782 | 0.0238 | No | ||
| 29 | TSHR | 2525 | 1.729 | 0.0216 | No | ||
| 30 | IL12A | 2616 | 1.689 | 0.0199 | No | ||
| 31 | SPINK2 | 2722 | 1.639 | 0.0172 | No | ||
| 32 | CDC42 | 2751 | 1.629 | 0.0187 | No | ||
| 33 | RUNX3 | 2761 | 1.626 | 0.0213 | No | ||
| 34 | HOXA9 | 2809 | 1.606 | 0.0217 | No | ||
| 35 | MINPP1 | 2812 | 1.606 | 0.0246 | No | ||
| 36 | CTBP2 | 3186 | 1.473 | 0.0071 | No | ||
| 37 | ANXA1 | 3289 | 1.437 | 0.0042 | No | ||
| 38 | RSBN1 | 3327 | 1.421 | 0.0048 | No | ||
| 39 | TTK | 3434 | 1.386 | 0.0017 | No | ||
| 40 | GAB2 | 3505 | 1.363 | 0.0004 | No | ||
| 41 | SPRY2 | 3611 | 1.332 | -0.0028 | No | ||
| 42 | EMP3 | 3815 | 1.272 | -0.0115 | No | ||
| 43 | DUSP3 | 3826 | 1.267 | -0.0097 | No | ||
| 44 | ST3GAL1 | 3848 | 1.259 | -0.0084 | No | ||
| 45 | ATP11B | 3880 | 1.246 | -0.0078 | No | ||
| 46 | CDKN2C | 3937 | 1.231 | -0.0086 | No | ||
| 47 | QKI | 3943 | 1.229 | -0.0065 | No | ||
| 48 | PLAGL1 | 3948 | 1.228 | -0.0045 | No | ||
| 49 | NPTX2 | 4092 | 1.188 | -0.0100 | No | ||
| 50 | HIST1H2BJ | 4097 | 1.187 | -0.0080 | No | ||
| 51 | ADH5 | 4195 | 1.159 | -0.0111 | No | ||
| 52 | CD44 | 4364 | 1.122 | -0.0182 | No | ||
| 53 | SCG2 | 4463 | 1.095 | -0.0215 | No | ||
| 54 | SELENBP1 | 4477 | 1.091 | -0.0201 | No | ||
| 55 | H1F0 | 4482 | 1.090 | -0.0183 | No | ||
| 56 | GUCY1A3 | 4615 | 1.058 | -0.0235 | No | ||
| 57 | TAF6L | 4911 | 0.989 | -0.0377 | No | ||
| 58 | TFPI | 5026 | 0.965 | -0.0421 | No | ||
| 59 | KLF10 | 5168 | 0.927 | -0.0480 | No | ||
| 60 | SLC2A3 | 5173 | 0.926 | -0.0465 | No | ||
| 61 | NKTR | 5220 | 0.912 | -0.0473 | No | ||
| 62 | CCL3 | 5302 | 0.891 | -0.0501 | No | ||
| 63 | MTMR6 | 5403 | 0.864 | -0.0539 | No | ||
| 64 | G0S2 | 5787 | 0.777 | -0.0732 | No | ||
| 65 | FTO | 5807 | 0.774 | -0.0728 | No | ||
| 66 | HIST1H2BL | 5881 | 0.758 | -0.0754 | No | ||
| 67 | IFI30 | 5905 | 0.754 | -0.0752 | No | ||
| 68 | LDLR | 5992 | 0.734 | -0.0785 | No | ||
| 69 | CAT | 6159 | 0.700 | -0.0863 | No | ||
| 70 | FGFR1 | 6252 | 0.677 | -0.0900 | No | ||
| 71 | DDX3Y | 6443 | 0.638 | -0.0991 | No | ||
| 72 | SC4MOL | 6647 | 0.599 | -0.1090 | No | ||
| 73 | PPP1R16B | 6895 | 0.549 | -0.1214 | No | ||
| 74 | PTGER4 | 6953 | 0.540 | -0.1235 | No | ||
| 75 | CXCL2 | 6990 | 0.532 | -0.1245 | No | ||
| 76 | HES1 | 7013 | 0.526 | -0.1247 | No | ||
| 77 | NOTCH3 | 7040 | 0.521 | -0.1252 | No | ||
| 78 | PRSS1 | 7353 | 0.454 | -0.1413 | No | ||
| 79 | PRKCA | 7407 | 0.444 | -0.1433 | No | ||
| 80 | ATP9A | 7447 | 0.435 | -0.1446 | No | ||
| 81 | JARID2 | 7512 | 0.423 | -0.1473 | No | ||
| 82 | MLC1 | 7537 | 0.417 | -0.1478 | No | ||
| 83 | RCBTB2 | 7840 | 0.357 | -0.1636 | No | ||
| 84 | TUBB2A | 7897 | 0.345 | -0.1660 | No | ||
| 85 | SERPINE2 | 8207 | 0.280 | -0.1822 | No | ||
| 86 | PSPH | 8227 | 0.276 | -0.1828 | No | ||
| 87 | FRMD4B | 8304 | 0.261 | -0.1864 | No | ||
| 88 | ALCAM | 8429 | 0.242 | -0.1927 | No | ||
| 89 | ATF3 | 8450 | 0.234 | -0.1933 | No | ||
| 90 | ST18 | 8648 | 0.194 | -0.2037 | No | ||
| 91 | PPBP | 9055 | 0.115 | -0.2255 | No | ||
| 92 | BHLHB2 | 9134 | 0.102 | -0.2296 | No | ||
| 93 | CXADR | 9147 | 0.099 | -0.2301 | No | ||
| 94 | LMO4 | 9148 | 0.099 | -0.2299 | No | ||
| 95 | CASP8 | 9176 | 0.093 | -0.2312 | No | ||
| 96 | ZBTB16 | 9297 | 0.072 | -0.2375 | No | ||
| 97 | WASF1 | 9376 | 0.053 | -0.2417 | No | ||
| 98 | TIPARP | 9409 | 0.047 | -0.2433 | No | ||
| 99 | TRH | 9423 | 0.044 | -0.2440 | No | ||
| 100 | IGJ | 9491 | 0.029 | -0.2476 | No | ||
| 101 | SKAP2 | 9521 | 0.020 | -0.2491 | No | ||
| 102 | SPOCK2 | 9636 | 0.002 | -0.2553 | No | ||
| 103 | PLK2 | 9646 | -0.001 | -0.2558 | No | ||
| 104 | RNF144 | 9928 | -0.056 | -0.2709 | No | ||
| 105 | RGS2 | 10067 | -0.085 | -0.2783 | No | ||
| 106 | HIST1H2AC | 10089 | -0.089 | -0.2792 | No | ||
| 107 | ETV5 | 10212 | -0.112 | -0.2857 | No | ||
| 108 | CTSH | 10506 | -0.169 | -0.3013 | No | ||
| 109 | ARID5B | 10661 | -0.206 | -0.3093 | No | ||
| 110 | SRPK2 | 10738 | -0.222 | -0.3130 | No | ||
| 111 | IRF5 | 10764 | -0.229 | -0.3139 | No | ||
| 112 | LAPTM4B | 11060 | -0.290 | -0.3294 | No | ||
| 113 | CRADD | 11312 | -0.342 | -0.3424 | No | ||
| 114 | SLC4A1 | 11565 | -0.397 | -0.3553 | No | ||
| 115 | S100A4 | 11583 | -0.401 | -0.3555 | No | ||
| 116 | LMO2 | 11761 | -0.443 | -0.3643 | No | ||
| 117 | FOS | 11810 | -0.455 | -0.3661 | No | ||
| 118 | IGLL1 | 12234 | -0.540 | -0.3881 | No | ||
| 119 | FOSB | 12305 | -0.556 | -0.3908 | No | ||
| 120 | ADFP | 12401 | -0.580 | -0.3949 | No | ||
| 121 | CD2 | 12514 | -0.609 | -0.3999 | No | ||
| 122 | FLT3 | 12882 | -0.693 | -0.4185 | No | ||
| 123 | IFRD1 | 12982 | -0.715 | -0.4225 | No | ||
| 124 | HOXA5 | 13292 | -0.800 | -0.4378 | No | ||
| 125 | CLEC11A | 13324 | -0.805 | -0.4380 | No | ||
| 126 | SPON1 | 13327 | -0.805 | -0.4366 | No | ||
| 127 | IDS | 13380 | -0.819 | -0.4379 | No | ||
| 128 | RB1CC1 | 13588 | -0.871 | -0.4476 | No | ||
| 129 | CDC7 | 13777 | -0.922 | -0.4561 | No | ||
| 130 | GABPB2 | 13912 | -0.961 | -0.4616 | No | ||
| 131 | AHR | 13940 | -0.971 | -0.4612 | No | ||
| 132 | PCDH9 | 13961 | -0.977 | -0.4605 | No | ||
| 133 | LRMP | 14007 | -0.988 | -0.4611 | No | ||
| 134 | PDLIM1 | 14048 | -0.997 | -0.4614 | No | ||
| 135 | RGS1 | 14130 | -1.022 | -0.4639 | No | ||
| 136 | IGFBP7 | 14240 | -1.055 | -0.4678 | No | ||
| 137 | CR2 | 14423 | -1.110 | -0.4757 | No | ||
| 138 | ALDH1A2 | 14476 | -1.127 | -0.4764 | No | ||
| 139 | CD200 | 14482 | -1.131 | -0.4746 | No | ||
| 140 | NID2 | 14522 | -1.141 | -0.4745 | No | ||
| 141 | LAT | 14531 | -1.143 | -0.4728 | No | ||
| 142 | FYB | 14563 | -1.152 | -0.4724 | No | ||
| 143 | CSDA | 14683 | -1.193 | -0.4766 | No | ||
| 144 | GARNL1 | 14827 | -1.247 | -0.4821 | No | ||
| 145 | BIRC3 | 14838 | -1.251 | -0.4803 | No | ||
| 146 | RORA | 14988 | -1.311 | -0.4859 | No | ||
| 147 | RYBP | 15007 | -1.320 | -0.4845 | No | ||
| 148 | SAP30 | 15127 | -1.369 | -0.4884 | No | ||
| 149 | SFPQ | 15284 | -1.436 | -0.4942 | No | ||
| 150 | HOXA10 | 15302 | -1.444 | -0.4924 | No | ||
| 151 | PRDX2 | 15315 | -1.450 | -0.4903 | No | ||
| 152 | IRF7 | 15384 | -1.486 | -0.4913 | No | ||
| 153 | ARF6 | 15400 | -1.497 | -0.4893 | No | ||
| 154 | CRIP1 | 15558 | -1.588 | -0.4949 | No | ||
| 155 | SLC35D1 | 15702 | -1.665 | -0.4995 | No | ||
| 156 | STK17B | 15768 | -1.700 | -0.4999 | No | ||
| 157 | ACTA1 | 15772 | -1.703 | -0.4969 | No | ||
| 158 | CLIC4 | 15822 | -1.729 | -0.4963 | No | ||
| 159 | MEF2A | 16038 | -1.869 | -0.5045 | No | ||
| 160 | PRDM2 | 16198 | -1.981 | -0.5095 | No | ||
| 161 | FHL1 | 16217 | -1.995 | -0.5067 | No | ||
| 162 | PMAIP1 | 16231 | -2.004 | -0.5037 | No | ||
| 163 | PTTG1IP | 16331 | -2.084 | -0.5052 | No | ||
| 164 | ITM2A | 16438 | -2.172 | -0.5069 | No | ||
| 165 | DUSP2 | 16622 | -2.321 | -0.5125 | Yes | ||
| 166 | MAL | 16626 | -2.323 | -0.5084 | Yes | ||
| 167 | EPHB6 | 16688 | -2.383 | -0.5072 | Yes | ||
| 168 | TRIB2 | 16696 | -2.391 | -0.5031 | Yes | ||
| 169 | PCTK2 | 16772 | -2.476 | -0.5026 | Yes | ||
| 170 | TAP1 | 16870 | -2.579 | -0.5031 | Yes | ||
| 171 | HPGD | 16871 | -2.580 | -0.4983 | Yes | ||
| 172 | TOB1 | 16926 | -2.641 | -0.4963 | Yes | ||
| 173 | UGCG | 17124 | -2.885 | -0.5016 | Yes | ||
| 174 | ETS2 | 17249 | -3.075 | -0.5026 | Yes | ||
| 175 | MPO | 17324 | -3.184 | -0.5007 | Yes | ||
| 176 | SELL | 17365 | -3.259 | -0.4968 | Yes | ||
| 177 | FXYD2 | 17448 | -3.408 | -0.4949 | Yes | ||
| 178 | HHEX | 17461 | -3.430 | -0.4891 | Yes | ||
| 179 | PIM2 | 17476 | -3.462 | -0.4834 | Yes | ||
| 180 | ARHGEF10 | 17534 | -3.573 | -0.4799 | Yes | ||
| 181 | CD97 | 17574 | -3.644 | -0.4752 | Yes | ||
| 182 | DUSP1 | 17723 | -3.944 | -0.4759 | Yes | ||
| 183 | S100A8 | 17813 | -4.140 | -0.4730 | Yes | ||
| 184 | RASGRP1 | 17841 | -4.206 | -0.4666 | Yes | ||
| 185 | RAP2B | 17931 | -4.484 | -0.4631 | Yes | ||
| 186 | ARID5A | 17950 | -4.542 | -0.4556 | Yes | ||
| 187 | SNF1LK | 17954 | -4.564 | -0.4473 | Yes | ||
| 188 | S100A9 | 17987 | -4.664 | -0.4403 | Yes | ||
| 189 | SOCS2 | 18077 | -4.952 | -0.4359 | Yes | ||
| 190 | DNTT | 18152 | -5.329 | -0.4300 | Yes | ||
| 191 | CCND2 | 18193 | -5.504 | -0.4219 | Yes | ||
| 192 | ALAS2 | 18249 | -5.792 | -0.4141 | Yes | ||
| 193 | BCL11A | 18279 | -6.014 | -0.4045 | Yes | ||
| 194 | IL6ST | 18347 | -6.519 | -0.3960 | Yes | ||
| 195 | CD69 | 18406 | -7.048 | -0.3860 | Yes | ||
| 196 | STAT1 | 18408 | -7.085 | -0.3728 | Yes | ||
| 197 | ISG20 | 18411 | -7.137 | -0.3596 | Yes | ||
| 198 | SLAMF1 | 18421 | -7.219 | -0.3467 | Yes | ||
| 199 | ARNTL | 18441 | -7.483 | -0.3338 | Yes | ||
| 200 | ITK | 18446 | -7.525 | -0.3199 | Yes | ||
| 201 | ANXA5 | 18461 | -7.790 | -0.3062 | Yes | ||
| 202 | CDKN1B | 18510 | -8.697 | -0.2926 | Yes | ||
| 203 | BCL6 | 18521 | -9.266 | -0.2758 | Yes | ||
| 204 | CD52 | 18538 | -9.945 | -0.2582 | Yes | ||
| 205 | ARL4C | 18547 | -10.194 | -0.2396 | Yes | ||
| 206 | DUSP6 | 18554 | -10.586 | -0.2202 | Yes | ||
| 207 | RAB6IP1 | 18567 | -12.306 | -0.1979 | Yes | ||
| 208 | TLE4 | 18570 | -12.734 | -0.1743 | Yes | ||
| 209 | CCL5 | 18571 | -12.889 | -0.1502 | Yes | ||
| 210 | ARMCX2 | 18572 | -13.045 | -0.1259 | Yes | ||
| 211 | S100A10 | 18577 | -13.802 | -0.1004 | Yes | ||
| 212 | MGAT5 | 18592 | -16.802 | -0.0698 | Yes | ||
| 213 | IL4R | 18594 | -17.159 | -0.0379 | Yes | ||
| 214 | ITGA6 | 18608 | -20.949 | 0.0004 | Yes |
