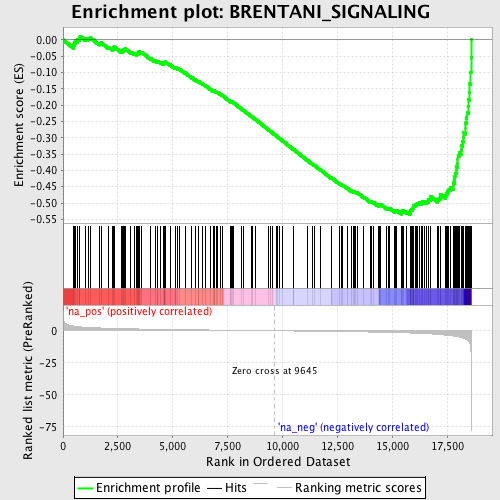
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | BRENTANI_SIGNALING |
| Enrichment Score (ES) | -0.5351005 |
| Normalized Enrichment Score (NES) | -1.9784764 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.12699246 |
| FWER p-Value | 0.644 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAPKAPK2 | 468 | 3.735 | -0.0156 | No | ||
| 2 | MAP3K7IP1 | 524 | 3.584 | -0.0093 | No | ||
| 3 | STAT5A | 573 | 3.481 | -0.0028 | No | ||
| 4 | ITGA9 | 658 | 3.265 | 0.0012 | No | ||
| 5 | TANK | 742 | 3.098 | 0.0048 | No | ||
| 6 | ERG | 768 | 3.062 | 0.0114 | No | ||
| 7 | TNFSF4 | 1019 | 2.729 | 0.0050 | No | ||
| 8 | IL12RB1 | 1136 | 2.605 | 0.0055 | No | ||
| 9 | TNFRSF10B | 1237 | 2.520 | 0.0066 | No | ||
| 10 | CRK | 1653 | 2.190 | -0.0102 | No | ||
| 11 | JAG2 | 1726 | 2.143 | -0.0085 | No | ||
| 12 | TYK2 | 2086 | 1.942 | -0.0229 | No | ||
| 13 | STAT6 | 2266 | 1.853 | -0.0277 | No | ||
| 14 | MPL | 2282 | 1.845 | -0.0237 | No | ||
| 15 | JAK3 | 2330 | 1.824 | -0.0215 | No | ||
| 16 | ABL1 | 2657 | 1.666 | -0.0348 | No | ||
| 17 | ITGA4 | 2710 | 1.643 | -0.0334 | No | ||
| 18 | HRAS | 2740 | 1.631 | -0.0307 | No | ||
| 19 | CXCL1 | 2793 | 1.613 | -0.0293 | No | ||
| 20 | NOTCH4 | 2832 | 1.599 | -0.0272 | No | ||
| 21 | PTCH2 | 3093 | 1.507 | -0.0374 | No | ||
| 22 | APC | 3246 | 1.454 | -0.0418 | No | ||
| 23 | TYRO3 | 3347 | 1.413 | -0.0435 | No | ||
| 24 | MAS1 | 3397 | 1.398 | -0.0425 | No | ||
| 25 | SOS1 | 3421 | 1.389 | -0.0402 | No | ||
| 26 | TTK | 3434 | 1.386 | -0.0372 | No | ||
| 27 | REL | 3458 | 1.377 | -0.0349 | No | ||
| 28 | WNT1 | 3594 | 1.337 | -0.0387 | No | ||
| 29 | WNT5A | 3996 | 1.215 | -0.0573 | No | ||
| 30 | DLG1 | 4188 | 1.161 | -0.0646 | No | ||
| 31 | MAP2K7 | 4298 | 1.135 | -0.0675 | No | ||
| 32 | BCR | 4306 | 1.134 | -0.0649 | No | ||
| 33 | RAN | 4431 | 1.102 | -0.0688 | No | ||
| 34 | IL1B | 4562 | 1.071 | -0.0730 | No | ||
| 35 | TNF | 4598 | 1.062 | -0.0722 | No | ||
| 36 | BIRC2 | 4618 | 1.057 | -0.0704 | No | ||
| 37 | MYD88 | 4630 | 1.054 | -0.0683 | No | ||
| 38 | NCK1 | 4671 | 1.044 | -0.0677 | No | ||
| 39 | MUSK | 4874 | 0.996 | -0.0761 | No | ||
| 40 | MAPK10 | 5114 | 0.942 | -0.0866 | No | ||
| 41 | INHA | 5121 | 0.939 | -0.0845 | No | ||
| 42 | ARHGAP1 | 5222 | 0.911 | -0.0875 | No | ||
| 43 | MAPKAPK5 | 5303 | 0.891 | -0.0895 | No | ||
| 44 | ARHGAP6 | 5558 | 0.831 | -0.1011 | No | ||
| 45 | MAP2K4 | 5861 | 0.762 | -0.1155 | No | ||
| 46 | ARHGEF12 | 6020 | 0.728 | -0.1222 | No | ||
| 47 | ITGAM | 6157 | 0.700 | -0.1277 | No | ||
| 48 | MAPK13 | 6168 | 0.698 | -0.1264 | No | ||
| 49 | ROCK1 | 6347 | 0.656 | -0.1344 | No | ||
| 50 | ITGA3 | 6494 | 0.629 | -0.1406 | No | ||
| 51 | GRB14 | 6729 | 0.586 | -0.1518 | No | ||
| 52 | SUFU | 6853 | 0.557 | -0.1570 | No | ||
| 53 | WNT4 | 6877 | 0.553 | -0.1568 | No | ||
| 54 | AKT1 | 6886 | 0.551 | -0.1558 | No | ||
| 55 | CXCL2 | 6990 | 0.532 | -0.1600 | No | ||
| 56 | NOTCH3 | 7040 | 0.521 | -0.1613 | No | ||
| 57 | IL13 | 7154 | 0.500 | -0.1661 | No | ||
| 58 | TNFSF13 | 7243 | 0.476 | -0.1696 | No | ||
| 59 | MCC | 7632 | 0.400 | -0.1896 | No | ||
| 60 | IGFBP3 | 7649 | 0.397 | -0.1895 | No | ||
| 61 | MAP2K2 | 7680 | 0.391 | -0.1901 | No | ||
| 62 | SMO | 7698 | 0.388 | -0.1900 | No | ||
| 63 | CAMK4 | 7761 | 0.376 | -0.1924 | No | ||
| 64 | ITGB7 | 8124 | 0.298 | -0.2112 | No | ||
| 65 | ITGB5 | 8213 | 0.279 | -0.2152 | No | ||
| 66 | IFNB1 | 8602 | 0.204 | -0.2357 | No | ||
| 67 | WNT2 | 8644 | 0.196 | -0.2375 | No | ||
| 68 | CALM1 | 8771 | 0.170 | -0.2438 | No | ||
| 69 | NRAS | 9380 | 0.052 | -0.2767 | No | ||
| 70 | MAP2K1 | 9431 | 0.042 | -0.2793 | No | ||
| 71 | ARHGEF16 | 9538 | 0.019 | -0.2850 | No | ||
| 72 | BTK | 9731 | -0.016 | -0.2953 | No | ||
| 73 | SKIL | 9772 | -0.022 | -0.2974 | No | ||
| 74 | YWHAE | 9858 | -0.041 | -0.3019 | No | ||
| 75 | MAPK4 | 10018 | -0.073 | -0.3104 | No | ||
| 76 | RASA1 | 10493 | -0.165 | -0.3356 | No | ||
| 77 | MAP2K3 | 10511 | -0.169 | -0.3361 | No | ||
| 78 | IGFBP1 | 11123 | -0.303 | -0.3684 | No | ||
| 79 | SRC | 11366 | -0.352 | -0.3806 | No | ||
| 80 | MAP3K4 | 11463 | -0.376 | -0.3848 | No | ||
| 81 | GRB7 | 11709 | -0.430 | -0.3970 | No | ||
| 82 | ITGAX | 12228 | -0.539 | -0.4237 | No | ||
| 83 | RAP1A | 12241 | -0.541 | -0.4229 | No | ||
| 84 | ITGB1 | 12596 | -0.629 | -0.4405 | No | ||
| 85 | ZAP70 | 12681 | -0.648 | -0.4433 | No | ||
| 86 | MAPK7 | 12748 | -0.663 | -0.4452 | No | ||
| 87 | IFNAR2 | 12965 | -0.712 | -0.4550 | No | ||
| 88 | ABL2 | 13138 | -0.759 | -0.4624 | No | ||
| 89 | ROS1 | 13239 | -0.787 | -0.4657 | No | ||
| 90 | CXCL12 | 13280 | -0.794 | -0.4658 | No | ||
| 91 | IRS1 | 13344 | -0.810 | -0.4671 | No | ||
| 92 | TNFRSF1B | 13395 | -0.822 | -0.4677 | No | ||
| 93 | JAK1 | 13695 | -0.897 | -0.4816 | No | ||
| 94 | NF1 | 14026 | -0.992 | -0.4969 | No | ||
| 95 | IFNAR1 | 14046 | -0.997 | -0.4953 | No | ||
| 96 | MAP2K5 | 14164 | -1.034 | -0.4989 | No | ||
| 97 | RALBP1 | 14395 | -1.102 | -0.5085 | No | ||
| 98 | PTPRK | 14407 | -1.106 | -0.5062 | No | ||
| 99 | JAG1 | 14425 | -1.110 | -0.5043 | No | ||
| 100 | ITGAV | 14464 | -1.125 | -0.5034 | No | ||
| 101 | IL15 | 14748 | -1.215 | -0.5156 | No | ||
| 102 | AXL | 14846 | -1.255 | -0.5175 | No | ||
| 103 | ILK | 14888 | -1.271 | -0.5164 | No | ||
| 104 | PIK3CG | 15103 | -1.358 | -0.5245 | No | ||
| 105 | IFNG | 15149 | -1.377 | -0.5233 | No | ||
| 106 | PTEN | 15194 | -1.398 | -0.5221 | No | ||
| 107 | MAPK12 | 15410 | -1.504 | -0.5298 | No | ||
| 108 | PIK3C3 | 15418 | -1.509 | -0.5263 | No | ||
| 109 | ATM | 15455 | -1.529 | -0.5242 | No | ||
| 110 | STAT5B | 15505 | -1.559 | -0.5228 | No | ||
| 111 | VIPR1 | 15669 | -1.649 | -0.5273 | No | ||
| 112 | AXIN2 | 15813 | -1.724 | -0.5306 | Yes | ||
| 113 | IL10 | 15830 | -1.735 | -0.5269 | Yes | ||
| 114 | IL2RB | 15844 | -1.743 | -0.5231 | Yes | ||
| 115 | ITGB2 | 15859 | -1.753 | -0.5193 | Yes | ||
| 116 | MAP3K5 | 15909 | -1.779 | -0.5173 | Yes | ||
| 117 | RAF1 | 15947 | -1.805 | -0.5146 | Yes | ||
| 118 | MAPK1 | 15982 | -1.834 | -0.5117 | Yes | ||
| 119 | ITGA7 | 15984 | -1.836 | -0.5069 | Yes | ||
| 120 | SYK | 16044 | -1.873 | -0.5052 | Yes | ||
| 121 | MAP3K14 | 16110 | -1.919 | -0.5038 | Yes | ||
| 122 | RELB | 16143 | -1.946 | -0.5004 | Yes | ||
| 123 | ITGB3 | 16223 | -2.000 | -0.4995 | Yes | ||
| 124 | SHC1 | 16319 | -2.078 | -0.4992 | Yes | ||
| 125 | SFN | 16365 | -2.109 | -0.4962 | Yes | ||
| 126 | MKNK1 | 16480 | -2.203 | -0.4966 | Yes | ||
| 127 | AKT2 | 16584 | -2.289 | -0.4962 | Yes | ||
| 128 | MAP4K4 | 16650 | -2.348 | -0.4936 | Yes | ||
| 129 | TRAF6 | 16659 | -2.353 | -0.4879 | Yes | ||
| 130 | RELA | 16736 | -2.440 | -0.4857 | Yes | ||
| 131 | GNB1 | 16756 | -2.461 | -0.4803 | Yes | ||
| 132 | IL18R1 | 17080 | -2.832 | -0.4904 | Yes | ||
| 133 | MAPK9 | 17129 | -2.896 | -0.4854 | Yes | ||
| 134 | SH3BP2 | 17187 | -2.977 | -0.4808 | Yes | ||
| 135 | TIAM1 | 17194 | -2.990 | -0.4733 | Yes | ||
| 136 | ITGA5 | 17440 | -3.396 | -0.4777 | Yes | ||
| 137 | BCL10 | 17457 | -3.421 | -0.4697 | Yes | ||
| 138 | ITGAL | 17513 | -3.538 | -0.4634 | Yes | ||
| 139 | VAV1 | 17585 | -3.659 | -0.4577 | Yes | ||
| 140 | MAP3K8 | 17644 | -3.789 | -0.4510 | Yes | ||
| 141 | MAP3K12 | 17778 | -4.065 | -0.4476 | Yes | ||
| 142 | IKBKG | 17782 | -4.070 | -0.4371 | Yes | ||
| 143 | ITGAE | 17831 | -4.185 | -0.4288 | Yes | ||
| 144 | FYN | 17854 | -4.233 | -0.4190 | Yes | ||
| 145 | IFNGR1 | 17866 | -4.284 | -0.4084 | Yes | ||
| 146 | MAPK11 | 17912 | -4.438 | -0.3992 | Yes | ||
| 147 | MAP2K6 | 17919 | -4.460 | -0.3879 | Yes | ||
| 148 | DGKQ | 17976 | -4.621 | -0.3789 | Yes | ||
| 149 | CRKL | 17994 | -4.686 | -0.3676 | Yes | ||
| 150 | STAT3 | 17998 | -4.700 | -0.3555 | Yes | ||
| 151 | MAP3K3 | 18061 | -4.882 | -0.3462 | Yes | ||
| 152 | ITGB4 | 18137 | -5.231 | -0.3366 | Yes | ||
| 153 | STAT2 | 18166 | -5.430 | -0.3239 | Yes | ||
| 154 | TNFRSF1A | 18199 | -5.538 | -0.3112 | Yes | ||
| 155 | TNFRSF14 | 18235 | -5.715 | -0.2982 | Yes | ||
| 156 | PDPK1 | 18265 | -5.919 | -0.2843 | Yes | ||
| 157 | IL6ST | 18347 | -6.519 | -0.2717 | Yes | ||
| 158 | AXIN1 | 18349 | -6.535 | -0.2547 | Yes | ||
| 159 | TRAF3 | 18385 | -6.833 | -0.2388 | Yes | ||
| 160 | STAT1 | 18408 | -7.085 | -0.2215 | Yes | ||
| 161 | TRAF1 | 18476 | -8.002 | -0.2043 | Yes | ||
| 162 | RGS16 | 18486 | -8.202 | -0.1834 | Yes | ||
| 163 | DGKA | 18533 | -9.632 | -0.1608 | Yes | ||
| 164 | PTK2B | 18539 | -9.957 | -0.1351 | Yes | ||
| 165 | TNFRSF4 | 18579 | -14.143 | -0.1003 | Yes | ||
| 166 | DGKG | 18597 | -18.226 | -0.0537 | Yes | ||
| 167 | ITGA6 | 18608 | -20.949 | 0.0004 | Yes |
