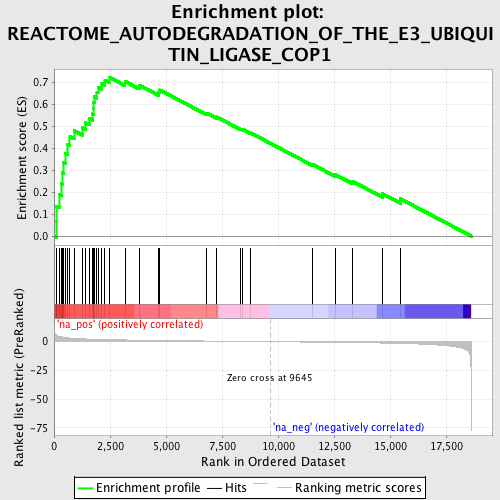
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_AUTODEGRADATION_OF_THE_E3_UBIQUITIN_LIGASE_COP1 |
| Enrichment Score (ES) | 0.7231534 |
| Normalized Enrichment Score (NES) | 2.4674418 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMB1 | 117 | 5.485 | 0.0651 | Yes | ||
| 2 | PSMC5 | 120 | 5.464 | 0.1361 | Yes | ||
| 3 | PSMB2 | 231 | 4.585 | 0.1899 | Yes | ||
| 4 | PSMB10 | 340 | 4.153 | 0.2382 | Yes | ||
| 5 | PSMD14 | 372 | 4.009 | 0.2887 | Yes | ||
| 6 | PSMD12 | 420 | 3.849 | 0.3363 | Yes | ||
| 7 | PSMD3 | 517 | 3.605 | 0.3781 | Yes | ||
| 8 | PSMD8 | 600 | 3.404 | 0.4180 | Yes | ||
| 9 | PSMC3 | 708 | 3.161 | 0.4534 | Yes | ||
| 10 | PSME3 | 901 | 2.859 | 0.4803 | Yes | ||
| 11 | PSMA4 | 1262 | 2.491 | 0.4933 | Yes | ||
| 12 | PSMC6 | 1396 | 2.364 | 0.5169 | Yes | ||
| 13 | PSMA7 | 1576 | 2.237 | 0.5364 | Yes | ||
| 14 | PSMC2 | 1712 | 2.153 | 0.5572 | Yes | ||
| 15 | PSMB6 | 1762 | 2.123 | 0.5822 | Yes | ||
| 16 | PSMB9 | 1773 | 2.117 | 0.6092 | Yes | ||
| 17 | TP53 | 1792 | 2.107 | 0.6357 | Yes | ||
| 18 | PSMA2 | 1913 | 2.034 | 0.6557 | Yes | ||
| 19 | PSMB5 | 1964 | 2.005 | 0.6791 | Yes | ||
| 20 | PSMA5 | 2135 | 1.923 | 0.6950 | Yes | ||
| 21 | PSMB3 | 2270 | 1.851 | 0.7119 | Yes | ||
| 22 | PSMD11 | 2485 | 1.748 | 0.7232 | Yes | ||
| 23 | PSMD7 | 3164 | 1.479 | 0.7059 | No | ||
| 24 | PSME1 | 3795 | 1.278 | 0.6887 | No | ||
| 25 | PSMA1 | 4659 | 1.048 | 0.6559 | No | ||
| 26 | PSMD9 | 4711 | 1.035 | 0.6666 | No | ||
| 27 | PSMD2 | 6824 | 0.564 | 0.5603 | No | ||
| 28 | PSMA6 | 7253 | 0.475 | 0.5434 | No | ||
| 29 | PSMC1 | 8340 | 0.255 | 0.4883 | No | ||
| 30 | PSMD10 | 8423 | 0.242 | 0.4870 | No | ||
| 31 | PSMD5 | 8760 | 0.172 | 0.4712 | No | ||
| 32 | PSMB4 | 11512 | -0.386 | 0.3282 | No | ||
| 33 | PSMD4 | 12572 | -0.622 | 0.2793 | No | ||
| 34 | PSMA3 | 13335 | -0.808 | 0.2488 | No | ||
| 35 | PSMC4 | 14658 | -1.186 | 0.1931 | No | ||
| 36 | ATM | 15455 | -1.529 | 0.1701 | No |
