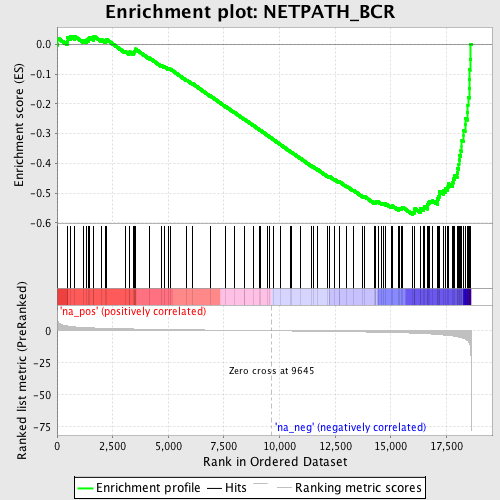
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.57160157 |
| Normalized Enrichment Score (NES) | -2.0413663 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06689931 |
| FWER p-Value | 0.238 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BAX | 38 | 6.993 | 0.0213 | No | ||
| 2 | CDK4 | 450 | 3.776 | 0.0117 | No | ||
| 3 | MAPKAPK2 | 468 | 3.735 | 0.0233 | No | ||
| 4 | RPS6 | 591 | 3.427 | 0.0281 | No | ||
| 5 | PTK2 | 791 | 3.028 | 0.0275 | No | ||
| 6 | RPS6KA1 | 1191 | 2.561 | 0.0145 | No | ||
| 7 | ITPR2 | 1328 | 2.420 | 0.0152 | No | ||
| 8 | CYCS | 1401 | 2.360 | 0.0192 | No | ||
| 9 | PIK3R2 | 1444 | 2.324 | 0.0247 | No | ||
| 10 | GRB2 | 1642 | 2.197 | 0.0214 | No | ||
| 11 | CRK | 1653 | 2.190 | 0.0282 | No | ||
| 12 | PTPRC | 1973 | 2.001 | 0.0176 | No | ||
| 13 | LYN | 2171 | 1.899 | 0.0133 | No | ||
| 14 | NFATC2 | 2224 | 1.873 | 0.0168 | No | ||
| 15 | PPP3R1 | 3054 | 1.521 | -0.0229 | No | ||
| 16 | CCNA2 | 3234 | 1.458 | -0.0277 | No | ||
| 17 | FCGR2B | 3274 | 1.441 | -0.0250 | No | ||
| 18 | SOS1 | 3421 | 1.389 | -0.0283 | No | ||
| 19 | REL | 3458 | 1.377 | -0.0256 | No | ||
| 20 | RAPGEF1 | 3484 | 1.369 | -0.0224 | No | ||
| 21 | GAB2 | 3505 | 1.363 | -0.0189 | No | ||
| 22 | GSK3A | 3517 | 1.358 | -0.0150 | No | ||
| 23 | PRKCD | 4150 | 1.173 | -0.0452 | No | ||
| 24 | NCK1 | 4671 | 1.044 | -0.0698 | No | ||
| 25 | SOS2 | 4831 | 1.008 | -0.0750 | No | ||
| 26 | MAP3K7 | 5000 | 0.970 | -0.0809 | No | ||
| 27 | DOK1 | 5087 | 0.949 | -0.0824 | No | ||
| 28 | PIK3R1 | 5830 | 0.769 | -0.1199 | No | ||
| 29 | HCLS1 | 6090 | 0.713 | -0.1315 | No | ||
| 30 | AKT1 | 6886 | 0.551 | -0.1726 | No | ||
| 31 | CDK7 | 7589 | 0.409 | -0.2092 | No | ||
| 32 | CD22 | 7961 | 0.334 | -0.2281 | No | ||
| 33 | ACTR2 | 8413 | 0.245 | -0.2517 | No | ||
| 34 | BLK | 8442 | 0.237 | -0.2524 | No | ||
| 35 | HCK | 8805 | 0.162 | -0.2715 | No | ||
| 36 | BCAR1 | 9088 | 0.110 | -0.2863 | No | ||
| 37 | CHUK | 9136 | 0.102 | -0.2885 | No | ||
| 38 | CD72 | 9464 | 0.035 | -0.3061 | No | ||
| 39 | LAT2 | 9562 | 0.015 | -0.3113 | No | ||
| 40 | BTK | 9731 | -0.016 | -0.3203 | No | ||
| 41 | MAPK4 | 10018 | -0.073 | -0.3355 | No | ||
| 42 | RASA1 | 10493 | -0.165 | -0.3606 | No | ||
| 43 | CD79A | 10539 | -0.174 | -0.3624 | No | ||
| 44 | CD19 | 10950 | -0.264 | -0.3837 | No | ||
| 45 | ACTR3 | 11419 | -0.366 | -0.4078 | No | ||
| 46 | DOK3 | 11511 | -0.386 | -0.4114 | No | ||
| 47 | MAPK3 | 11722 | -0.435 | -0.4213 | No | ||
| 48 | PRKCE | 12144 | -0.521 | -0.4423 | No | ||
| 49 | IKBKB | 12258 | -0.546 | -0.4466 | No | ||
| 50 | RASGRP3 | 12262 | -0.547 | -0.4449 | No | ||
| 51 | BLNK | 12466 | -0.598 | -0.4539 | No | ||
| 52 | ZAP70 | 12681 | -0.648 | -0.4633 | No | ||
| 53 | CDK6 | 12685 | -0.649 | -0.4613 | No | ||
| 54 | CCNE1 | 13017 | -0.724 | -0.4768 | No | ||
| 55 | PIK3AP1 | 13310 | -0.802 | -0.4899 | No | ||
| 56 | HDAC5 | 13739 | -0.908 | -0.5100 | No | ||
| 57 | PTPN6 | 13818 | -0.932 | -0.5111 | No | ||
| 58 | JUN | 14245 | -1.057 | -0.5306 | No | ||
| 59 | CBLB | 14325 | -1.083 | -0.5312 | No | ||
| 60 | PLCG1 | 14331 | -1.084 | -0.5279 | No | ||
| 61 | CR2 | 14423 | -1.110 | -0.5291 | No | ||
| 62 | PPP3CA | 14583 | -1.160 | -0.5338 | No | ||
| 63 | RPS6KB1 | 14689 | -1.195 | -0.5355 | No | ||
| 64 | CSK | 14774 | -1.225 | -0.5359 | No | ||
| 65 | MAPK8 | 15028 | -1.330 | -0.5451 | No | ||
| 66 | PTPN18 | 15062 | -1.343 | -0.5424 | No | ||
| 67 | WAS | 15336 | -1.459 | -0.5523 | No | ||
| 68 | MAPK14 | 15392 | -1.490 | -0.5503 | No | ||
| 69 | MAP4K1 | 15494 | -1.551 | -0.5506 | No | ||
| 70 | DAPP1 | 15522 | -1.570 | -0.5468 | No | ||
| 71 | MAPK1 | 15982 | -1.834 | -0.5655 | Yes | ||
| 72 | SYK | 16044 | -1.873 | -0.5625 | Yes | ||
| 73 | CASP9 | 16070 | -1.890 | -0.5575 | Yes | ||
| 74 | CD81 | 16082 | -1.898 | -0.5518 | Yes | ||
| 75 | SHC1 | 16319 | -2.078 | -0.5576 | Yes | ||
| 76 | PTPN11 | 16328 | -2.083 | -0.5511 | Yes | ||
| 77 | PRKD1 | 16464 | -2.190 | -0.5511 | Yes | ||
| 78 | CCND3 | 16501 | -2.216 | -0.5456 | Yes | ||
| 79 | RB1 | 16644 | -2.339 | -0.5455 | Yes | ||
| 80 | ATF2 | 16651 | -2.349 | -0.5379 | Yes | ||
| 81 | VAV2 | 16677 | -2.369 | -0.5314 | Yes | ||
| 82 | RELA | 16736 | -2.440 | -0.5264 | Yes | ||
| 83 | LCK | 16851 | -2.559 | -0.5240 | Yes | ||
| 84 | CDK2 | 17094 | -2.849 | -0.5275 | Yes | ||
| 85 | BCL2L11 | 17107 | -2.863 | -0.5186 | Yes | ||
| 86 | GSK3B | 17139 | -2.906 | -0.5106 | Yes | ||
| 87 | SH3BP2 | 17187 | -2.977 | -0.5032 | Yes | ||
| 88 | CREB1 | 17191 | -2.984 | -0.4933 | Yes | ||
| 89 | PRKCQ | 17373 | -3.270 | -0.4922 | Yes | ||
| 90 | BCL10 | 17457 | -3.421 | -0.4853 | Yes | ||
| 91 | NFKBIA | 17567 | -3.627 | -0.4790 | Yes | ||
| 92 | VAV1 | 17585 | -3.659 | -0.4677 | Yes | ||
| 93 | IKBKG | 17782 | -4.070 | -0.4647 | Yes | ||
| 94 | GTF2I | 17803 | -4.112 | -0.4520 | Yes | ||
| 95 | FYN | 17854 | -4.233 | -0.4406 | Yes | ||
| 96 | LCP2 | 17979 | -4.636 | -0.4318 | Yes | ||
| 97 | CRKL | 17994 | -4.686 | -0.4169 | Yes | ||
| 98 | NFATC1 | 18056 | -4.862 | -0.4040 | Yes | ||
| 99 | PDK2 | 18065 | -4.903 | -0.3880 | Yes | ||
| 100 | NFATC3 | 18098 | -5.049 | -0.3729 | Yes | ||
| 101 | CBL | 18150 | -5.305 | -0.3579 | Yes | ||
| 102 | CCND2 | 18193 | -5.504 | -0.3418 | Yes | ||
| 103 | CARD11 | 18195 | -5.524 | -0.3234 | Yes | ||
| 104 | PDPK1 | 18265 | -5.919 | -0.3073 | Yes | ||
| 105 | PIP5K1C | 18268 | -5.934 | -0.2876 | Yes | ||
| 106 | CD5 | 18337 | -6.400 | -0.2699 | Yes | ||
| 107 | CD79B | 18344 | -6.466 | -0.2486 | Yes | ||
| 108 | ITK | 18446 | -7.525 | -0.2289 | Yes | ||
| 109 | CASP7 | 18463 | -7.844 | -0.2036 | Yes | ||
| 110 | NEDD9 | 18470 | -7.962 | -0.1773 | Yes | ||
| 111 | BCL6 | 18521 | -9.266 | -0.1490 | Yes | ||
| 112 | BCL2 | 18530 | -9.542 | -0.1175 | Yes | ||
| 113 | PTK2B | 18539 | -9.957 | -0.0847 | Yes | ||
| 114 | GAB1 | 18560 | -10.871 | -0.0495 | Yes | ||
| 115 | TEC | 18587 | -15.689 | 0.0016 | Yes |
