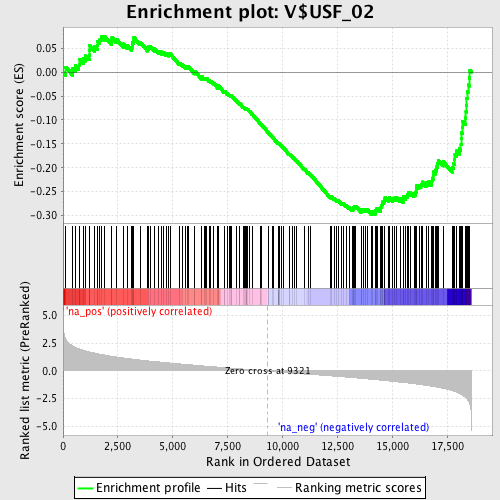
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$USF_02 |
| Enrichment Score (ES) | -0.2977954 |
| Normalized Enrichment Score (NES) | -1.5108526 |
| Nominal p-value | 0.0025316456 |
| FDR q-value | 0.20081797 |
| FWER p-Value | 0.967 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF4E | 124 | 2.878 | 0.0106 | No | ||
| 2 | COQ4 | 441 | 2.242 | 0.0070 | No | ||
| 3 | PTGES2 | 563 | 2.131 | 0.0133 | No | ||
| 4 | RNF128 | 724 | 1.989 | 0.0167 | No | ||
| 5 | AP1S2 | 763 | 1.959 | 0.0264 | No | ||
| 6 | BDNF | 938 | 1.854 | 0.0282 | No | ||
| 7 | TAC1 | 1033 | 1.798 | 0.0339 | No | ||
| 8 | ZFYVE26 | 1191 | 1.716 | 0.0358 | No | ||
| 9 | HNRPH2 | 1202 | 1.712 | 0.0456 | No | ||
| 10 | SOX10 | 1210 | 1.709 | 0.0555 | No | ||
| 11 | DUSP7 | 1433 | 1.619 | 0.0532 | No | ||
| 12 | SESN3 | 1553 | 1.572 | 0.0563 | No | ||
| 13 | HIVEP1 | 1575 | 1.561 | 0.0645 | No | ||
| 14 | ARMET | 1678 | 1.519 | 0.0682 | No | ||
| 15 | DUSP8 | 1730 | 1.500 | 0.0745 | No | ||
| 16 | IRS4 | 1877 | 1.444 | 0.0753 | No | ||
| 17 | SLC39A5 | 2192 | 1.332 | 0.0663 | No | ||
| 18 | SLC12A6 | 2226 | 1.322 | 0.0725 | No | ||
| 19 | GLA | 2431 | 1.261 | 0.0690 | No | ||
| 20 | UMPS | 2749 | 1.176 | 0.0589 | No | ||
| 21 | OPRS1 | 2930 | 1.127 | 0.0559 | No | ||
| 22 | SEC23IP | 3124 | 1.077 | 0.0520 | No | ||
| 23 | RAB9A | 3157 | 1.068 | 0.0567 | No | ||
| 24 | MCAM | 3170 | 1.067 | 0.0625 | No | ||
| 25 | TESK2 | 3218 | 1.057 | 0.0663 | No | ||
| 26 | C4A | 3223 | 1.055 | 0.0725 | No | ||
| 27 | NRIP3 | 3514 | 0.987 | 0.0627 | No | ||
| 28 | NEUROD1 | 3859 | 0.909 | 0.0495 | No | ||
| 29 | FGF14 | 3897 | 0.900 | 0.0529 | No | ||
| 30 | SEMA3A | 3975 | 0.884 | 0.0541 | No | ||
| 31 | CCAR1 | 4166 | 0.851 | 0.0489 | No | ||
| 32 | LAMA3 | 4351 | 0.815 | 0.0439 | No | ||
| 33 | ATF4 | 4469 | 0.794 | 0.0423 | No | ||
| 34 | GZMK | 4586 | 0.771 | 0.0407 | No | ||
| 35 | ISGF3G | 4702 | 0.748 | 0.0389 | No | ||
| 36 | MTCH2 | 4803 | 0.730 | 0.0379 | No | ||
| 37 | HOXB7 | 4879 | 0.717 | 0.0382 | No | ||
| 38 | RPS2 | 5310 | 0.641 | 0.0187 | No | ||
| 39 | ILF3 | 5441 | 0.616 | 0.0154 | No | ||
| 40 | BRDT | 5578 | 0.591 | 0.0116 | No | ||
| 41 | NDUFA7 | 5657 | 0.578 | 0.0109 | No | ||
| 42 | MRPL27 | 5692 | 0.574 | 0.0125 | No | ||
| 43 | UROD | 5997 | 0.523 | -0.0009 | No | ||
| 44 | OMG | 5999 | 0.523 | 0.0022 | No | ||
| 45 | PRPS1 | 6317 | 0.469 | -0.0121 | No | ||
| 46 | MAP4K1 | 6319 | 0.468 | -0.0094 | No | ||
| 47 | SLC1A7 | 6452 | 0.446 | -0.0138 | No | ||
| 48 | ARPP-19 | 6481 | 0.443 | -0.0127 | No | ||
| 49 | VNN1 | 6548 | 0.432 | -0.0136 | No | ||
| 50 | NFX1 | 6649 | 0.416 | -0.0165 | No | ||
| 51 | DHX35 | 6734 | 0.402 | -0.0187 | No | ||
| 52 | CTNS | 6853 | 0.381 | -0.0228 | No | ||
| 53 | PLS3 | 7058 | 0.349 | -0.0317 | No | ||
| 54 | CDKN2C | 7076 | 0.347 | -0.0306 | No | ||
| 55 | COPZ1 | 7077 | 0.347 | -0.0285 | No | ||
| 56 | UBE2B | 7349 | 0.300 | -0.0414 | No | ||
| 57 | STK16 | 7358 | 0.299 | -0.0400 | No | ||
| 58 | STC2 | 7478 | 0.281 | -0.0447 | No | ||
| 59 | EIF3S10 | 7573 | 0.267 | -0.0482 | No | ||
| 60 | CREB5 | 7611 | 0.260 | -0.0487 | No | ||
| 61 | RPS28 | 7659 | 0.255 | -0.0497 | No | ||
| 62 | DVL2 | 7665 | 0.254 | -0.0484 | No | ||
| 63 | HNRPD | 7887 | 0.223 | -0.0591 | No | ||
| 64 | FEN1 | 8037 | 0.197 | -0.0660 | No | ||
| 65 | ELAVL3 | 8050 | 0.196 | -0.0654 | No | ||
| 66 | CALR | 8226 | 0.171 | -0.0739 | No | ||
| 67 | DDIT3 | 8277 | 0.163 | -0.0756 | No | ||
| 68 | TAF6L | 8292 | 0.161 | -0.0754 | No | ||
| 69 | MAX | 8341 | 0.151 | -0.0771 | No | ||
| 70 | PIK3R1 | 8356 | 0.150 | -0.0769 | No | ||
| 71 | SUPV3L1 | 8367 | 0.148 | -0.0766 | No | ||
| 72 | EGLN2 | 8407 | 0.142 | -0.0779 | No | ||
| 73 | RHOQ | 8490 | 0.129 | -0.0815 | No | ||
| 74 | BMP6 | 8615 | 0.106 | -0.0876 | No | ||
| 75 | RFC1 | 8994 | 0.049 | -0.1078 | No | ||
| 76 | HOXD10 | 9005 | 0.048 | -0.1081 | No | ||
| 77 | CARKL | 9032 | 0.043 | -0.1092 | No | ||
| 78 | HOXB5 | 9351 | -0.006 | -0.1264 | No | ||
| 79 | ATP6V1A | 9366 | -0.008 | -0.1271 | No | ||
| 80 | LIN28 | 9535 | -0.032 | -0.1361 | No | ||
| 81 | PFKFB1 | 9574 | -0.038 | -0.1379 | No | ||
| 82 | FZD6 | 9803 | -0.081 | -0.1498 | No | ||
| 83 | TOP3A | 9808 | -0.082 | -0.1495 | No | ||
| 84 | GNB2 | 9823 | -0.085 | -0.1497 | No | ||
| 85 | CAPN6 | 9831 | -0.086 | -0.1496 | No | ||
| 86 | RUNX1 | 9839 | -0.087 | -0.1494 | No | ||
| 87 | UBXD2 | 9843 | -0.088 | -0.1491 | No | ||
| 88 | XPO1 | 9942 | -0.106 | -0.1537 | No | ||
| 89 | PPARGC1A | 10033 | -0.121 | -0.1579 | No | ||
| 90 | PIP5K1A | 10312 | -0.158 | -0.1720 | No | ||
| 91 | PICALM | 10336 | -0.162 | -0.1723 | No | ||
| 92 | TOM1L2 | 10432 | -0.177 | -0.1764 | No | ||
| 93 | HOXC5 | 10534 | -0.191 | -0.1807 | No | ||
| 94 | DNAJB5 | 10626 | -0.206 | -0.1844 | No | ||
| 95 | SLC23A2 | 10991 | -0.267 | -0.2025 | No | ||
| 96 | AATF | 11162 | -0.293 | -0.2100 | No | ||
| 97 | SIRT1 | 11267 | -0.307 | -0.2138 | No | ||
| 98 | SUMF1 | 12184 | -0.438 | -0.2608 | No | ||
| 99 | GPR65 | 12218 | -0.443 | -0.2599 | No | ||
| 100 | SQSTM1 | 12347 | -0.459 | -0.2641 | No | ||
| 101 | BTBD14B | 12479 | -0.479 | -0.2683 | No | ||
| 102 | CPT1A | 12538 | -0.489 | -0.2685 | No | ||
| 103 | TOPORS | 12707 | -0.517 | -0.2745 | No | ||
| 104 | HOXA11 | 12763 | -0.524 | -0.2743 | No | ||
| 105 | GABARAP | 12894 | -0.548 | -0.2781 | No | ||
| 106 | GTF2H1 | 13034 | -0.570 | -0.2822 | No | ||
| 107 | SLC17A2 | 13177 | -0.590 | -0.2863 | No | ||
| 108 | BAX | 13218 | -0.596 | -0.2849 | No | ||
| 109 | DNMT3A | 13247 | -0.602 | -0.2828 | No | ||
| 110 | ATP6V1C1 | 13283 | -0.610 | -0.2810 | No | ||
| 111 | ASNA1 | 13341 | -0.620 | -0.2803 | No | ||
| 112 | TCERG1 | 13581 | -0.663 | -0.2893 | No | ||
| 113 | AKAP12 | 13605 | -0.666 | -0.2865 | No | ||
| 114 | UBE2L6 | 13709 | -0.685 | -0.2880 | No | ||
| 115 | NPTX1 | 13761 | -0.694 | -0.2865 | No | ||
| 116 | PTMA | 13857 | -0.712 | -0.2874 | No | ||
| 117 | TYR | 14045 | -0.742 | -0.2931 | No | ||
| 118 | CLCN5 | 14102 | -0.752 | -0.2916 | No | ||
| 119 | NIT1 | 14218 | -0.776 | -0.2931 | Yes | ||
| 120 | POGK | 14248 | -0.783 | -0.2900 | Yes | ||
| 121 | SMCR8 | 14265 | -0.786 | -0.2861 | Yes | ||
| 122 | CD164 | 14336 | -0.800 | -0.2851 | Yes | ||
| 123 | TIMM8A | 14473 | -0.825 | -0.2874 | Yes | ||
| 124 | APLN | 14483 | -0.827 | -0.2829 | Yes | ||
| 125 | KCNE4 | 14500 | -0.831 | -0.2788 | Yes | ||
| 126 | CRLF1 | 14535 | -0.837 | -0.2756 | Yes | ||
| 127 | GGN | 14555 | -0.841 | -0.2715 | Yes | ||
| 128 | FMR1 | 14625 | -0.854 | -0.2701 | Yes | ||
| 129 | COMMD8 | 14657 | -0.861 | -0.2666 | Yes | ||
| 130 | H2AFY | 14669 | -0.863 | -0.2620 | Yes | ||
| 131 | SLC31A2 | 14816 | -0.895 | -0.2645 | Yes | ||
| 132 | LHFP | 14883 | -0.909 | -0.2626 | Yes | ||
| 133 | CYR61 | 15029 | -0.939 | -0.2648 | Yes | ||
| 134 | UBE4B | 15107 | -0.954 | -0.2632 | Yes | ||
| 135 | NCOA6 | 15189 | -0.969 | -0.2618 | Yes | ||
| 136 | HNRPA1 | 15361 | -1.006 | -0.2650 | Yes | ||
| 137 | LRP5 | 15497 | -1.034 | -0.2661 | Yes | ||
| 138 | TGFB2 | 15506 | -1.035 | -0.2602 | Yes | ||
| 139 | TCF12 | 15625 | -1.060 | -0.2602 | Yes | ||
| 140 | DDX3X | 15680 | -1.071 | -0.2567 | Yes | ||
| 141 | VPS16 | 15719 | -1.080 | -0.2523 | Yes | ||
| 142 | FOXD3 | 15840 | -1.109 | -0.2521 | Yes | ||
| 143 | NPM1 | 15993 | -1.153 | -0.2534 | Yes | ||
| 144 | MTHFD1 | 16076 | -1.172 | -0.2507 | Yes | ||
| 145 | GRN | 16091 | -1.175 | -0.2444 | Yes | ||
| 146 | MAT2A | 16104 | -1.177 | -0.2380 | Yes | ||
| 147 | RAB3IL1 | 16222 | -1.208 | -0.2370 | Yes | ||
| 148 | UBXD3 | 16312 | -1.236 | -0.2344 | Yes | ||
| 149 | UTP14A | 16367 | -1.248 | -0.2298 | Yes | ||
| 150 | CBX5 | 16548 | -1.294 | -0.2317 | Yes | ||
| 151 | PABPC1 | 16657 | -1.327 | -0.2296 | Yes | ||
| 152 | CLN3 | 16801 | -1.371 | -0.2291 | Yes | ||
| 153 | COMMD3 | 16815 | -1.377 | -0.2214 | Yes | ||
| 154 | SYNPO2L | 16889 | -1.400 | -0.2170 | Yes | ||
| 155 | CHD4 | 16895 | -1.401 | -0.2088 | Yes | ||
| 156 | ATF7IP | 16980 | -1.430 | -0.2047 | Yes | ||
| 157 | ALDH6A1 | 17024 | -1.445 | -0.1983 | Yes | ||
| 158 | RHEBL1 | 17052 | -1.457 | -0.1910 | Yes | ||
| 159 | ZIC3 | 17086 | -1.469 | -0.1839 | Yes | ||
| 160 | PPM1A | 17318 | -1.569 | -0.1870 | Yes | ||
| 161 | BLNK | 17729 | -1.762 | -0.1986 | Yes | ||
| 162 | DCTN4 | 17805 | -1.806 | -0.1917 | Yes | ||
| 163 | EIF4B | 17836 | -1.829 | -0.1823 | Yes | ||
| 164 | SNX8 | 17859 | -1.845 | -0.1724 | Yes | ||
| 165 | LAMC2 | 17930 | -1.892 | -0.1647 | Yes | ||
| 166 | RAI14 | 18060 | -2.013 | -0.1596 | Yes | ||
| 167 | MDM1 | 18133 | -2.082 | -0.1509 | Yes | ||
| 168 | GPX1 | 18151 | -2.105 | -0.1391 | Yes | ||
| 169 | ETV1 | 18164 | -2.117 | -0.1270 | Yes | ||
| 170 | NUDC | 18192 | -2.156 | -0.1155 | Yes | ||
| 171 | TNRC15 | 18202 | -2.165 | -0.1029 | Yes | ||
| 172 | KBTBD2 | 18334 | -2.357 | -0.0958 | Yes | ||
| 173 | PFDN2 | 18360 | -2.392 | -0.0827 | Yes | ||
| 174 | PER1 | 18385 | -2.454 | -0.0692 | Yes | ||
| 175 | SSR1 | 18396 | -2.475 | -0.0548 | Yes | ||
| 176 | VLDLR | 18420 | -2.522 | -0.0408 | Yes | ||
| 177 | GTF2A1 | 18458 | -2.667 | -0.0267 | Yes | ||
| 178 | DUSP1 | 18504 | -2.834 | -0.0121 | Yes | ||
| 179 | EPC1 | 18540 | -2.994 | 0.0041 | Yes |