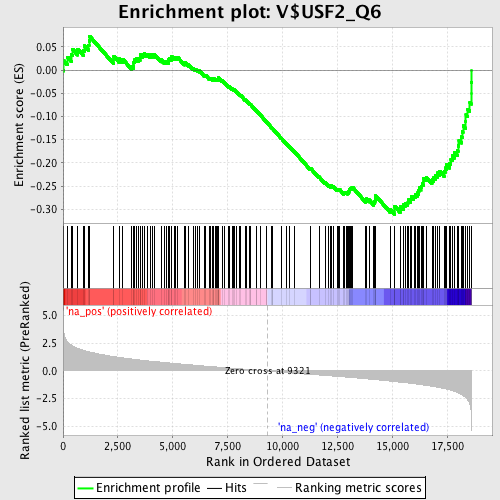
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$USF2_Q6 |
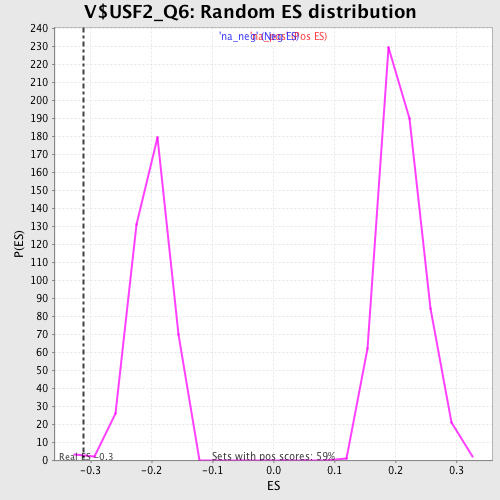
| Enrichment Score (ES) | -0.31140667 |
| Normalized Enrichment Score (NES) | -1.549786 |
| Nominal p-value | 0.00729927 |
| FDR q-value | 0.23675685 |
| FWER p-Value | 0.904 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BCL9L | 37 | 3.427 | 0.0200 | No | ||
| 2 | ZZZ3 | 197 | 2.635 | 0.0282 | No | ||
| 3 | IL15RA | 373 | 2.337 | 0.0337 | No | ||
| 4 | DPAGT1 | 434 | 2.260 | 0.0450 | No | ||
| 5 | TMEM24 | 666 | 2.025 | 0.0454 | No | ||
| 6 | BDNF | 938 | 1.854 | 0.0426 | No | ||
| 7 | RUSC1 | 974 | 1.834 | 0.0525 | No | ||
| 8 | TRIM3 | 1159 | 1.735 | 0.0536 | No | ||
| 9 | PTPRF | 1183 | 1.723 | 0.0634 | No | ||
| 10 | LRP8 | 1204 | 1.711 | 0.0733 | No | ||
| 11 | TRIM46 | 2305 | 1.298 | 0.0220 | No | ||
| 12 | RASGRP2 | 2310 | 1.295 | 0.0301 | No | ||
| 13 | ATF1 | 2561 | 1.224 | 0.0244 | No | ||
| 14 | NR0B2 | 2727 | 1.183 | 0.0230 | No | ||
| 15 | SEC23IP | 3124 | 1.077 | 0.0085 | No | ||
| 16 | FXYD2 | 3213 | 1.058 | 0.0105 | No | ||
| 17 | TESK2 | 3218 | 1.057 | 0.0170 | No | ||
| 18 | RAD9A | 3250 | 1.048 | 0.0221 | No | ||
| 19 | LDHA | 3329 | 1.029 | 0.0244 | No | ||
| 20 | RFX5 | 3443 | 1.004 | 0.0248 | No | ||
| 21 | ILK | 3507 | 0.989 | 0.0277 | No | ||
| 22 | NRIP3 | 3514 | 0.987 | 0.0337 | No | ||
| 23 | USP2 | 3631 | 0.960 | 0.0335 | No | ||
| 24 | TIAL1 | 3693 | 0.948 | 0.0363 | No | ||
| 25 | RARG | 3838 | 0.915 | 0.0344 | No | ||
| 26 | HMGN2 | 3971 | 0.885 | 0.0329 | No | ||
| 27 | B3GALT6 | 4061 | 0.868 | 0.0336 | No | ||
| 28 | CCAR1 | 4166 | 0.851 | 0.0334 | No | ||
| 29 | CENTB5 | 4462 | 0.795 | 0.0226 | No | ||
| 30 | B3GALT2 | 4635 | 0.760 | 0.0181 | No | ||
| 31 | PFKFB3 | 4698 | 0.748 | 0.0195 | No | ||
| 32 | MTCH2 | 4803 | 0.730 | 0.0186 | No | ||
| 33 | CFL1 | 4817 | 0.729 | 0.0226 | No | ||
| 34 | HOXC13 | 4844 | 0.724 | 0.0258 | No | ||
| 35 | NET1 | 4927 | 0.709 | 0.0259 | No | ||
| 36 | KRTCAP2 | 4948 | 0.705 | 0.0293 | No | ||
| 37 | TDRD1 | 5083 | 0.681 | 0.0264 | No | ||
| 38 | CHRM1 | 5138 | 0.669 | 0.0278 | No | ||
| 39 | COPS7A | 5229 | 0.653 | 0.0271 | No | ||
| 40 | RGL1 | 5528 | 0.599 | 0.0148 | No | ||
| 41 | BRDT | 5578 | 0.591 | 0.0159 | No | ||
| 42 | PLA2G4A | 5724 | 0.569 | 0.0117 | No | ||
| 43 | HOXC12 | 5932 | 0.534 | 0.0039 | No | ||
| 44 | BLOC1S1 | 6035 | 0.516 | 0.0017 | No | ||
| 45 | AMPD2 | 6127 | 0.500 | -0.0000 | No | ||
| 46 | IPO13 | 6210 | 0.487 | -0.0014 | No | ||
| 47 | SLC1A7 | 6452 | 0.446 | -0.0116 | No | ||
| 48 | CGN | 6490 | 0.441 | -0.0107 | No | ||
| 49 | POLR3C | 6682 | 0.410 | -0.0185 | No | ||
| 50 | CDC14A | 6712 | 0.405 | -0.0174 | No | ||
| 51 | BHLHB3 | 6798 | 0.390 | -0.0195 | No | ||
| 52 | PPP1R3C | 6854 | 0.381 | -0.0201 | No | ||
| 53 | PSEN2 | 6855 | 0.381 | -0.0176 | No | ||
| 54 | PA2G4 | 6935 | 0.368 | -0.0196 | No | ||
| 55 | ARL2 | 6998 | 0.359 | -0.0206 | No | ||
| 56 | HPCAL4 | 7016 | 0.356 | -0.0193 | No | ||
| 57 | CDKN2C | 7076 | 0.347 | -0.0202 | No | ||
| 58 | COPZ1 | 7077 | 0.347 | -0.0180 | No | ||
| 59 | PPRC1 | 7084 | 0.345 | -0.0161 | No | ||
| 60 | SCYL1 | 7247 | 0.318 | -0.0229 | No | ||
| 61 | AVPI1 | 7362 | 0.298 | -0.0271 | No | ||
| 62 | CNNM1 | 7551 | 0.271 | -0.0356 | No | ||
| 63 | EIF3S10 | 7573 | 0.267 | -0.0350 | No | ||
| 64 | LZTS2 | 7703 | 0.247 | -0.0404 | No | ||
| 65 | B4GALT2 | 7753 | 0.241 | -0.0415 | No | ||
| 66 | ARL3 | 7796 | 0.236 | -0.0423 | No | ||
| 67 | GUCY1A2 | 7916 | 0.218 | -0.0474 | No | ||
| 68 | FEN1 | 8037 | 0.197 | -0.0526 | No | ||
| 69 | ESRRA | 8099 | 0.189 | -0.0547 | No | ||
| 70 | TAF6L | 8292 | 0.161 | -0.0641 | No | ||
| 71 | LHX9 | 8344 | 0.151 | -0.0659 | No | ||
| 72 | IPO7 | 8474 | 0.131 | -0.0720 | No | ||
| 73 | FOSL1 | 8533 | 0.121 | -0.0744 | No | ||
| 74 | APOA5 | 8547 | 0.118 | -0.0743 | No | ||
| 75 | SLCO1C1 | 8807 | 0.076 | -0.0879 | No | ||
| 76 | AP3M1 | 8818 | 0.075 | -0.0879 | No | ||
| 77 | PITX3 | 9001 | 0.048 | -0.0975 | No | ||
| 78 | OPRD1 | 9257 | 0.011 | -0.1112 | No | ||
| 79 | BCL9 | 9270 | 0.008 | -0.1118 | No | ||
| 80 | TEX12 | 9490 | -0.026 | -0.1235 | No | ||
| 81 | ATP5F1 | 9532 | -0.032 | -0.1256 | No | ||
| 82 | LIN28 | 9535 | -0.032 | -0.1255 | No | ||
| 83 | RTN4RL2 | 9958 | -0.109 | -0.1476 | No | ||
| 84 | AGMAT | 10188 | -0.142 | -0.1591 | No | ||
| 85 | PICALM | 10336 | -0.162 | -0.1661 | No | ||
| 86 | HOXC5 | 10534 | -0.191 | -0.1755 | No | ||
| 87 | SIRT1 | 11267 | -0.307 | -0.2132 | No | ||
| 88 | HTATIP | 11272 | -0.308 | -0.2114 | No | ||
| 89 | ATAD3A | 11665 | -0.360 | -0.2304 | No | ||
| 90 | FCHSD2 | 11939 | -0.400 | -0.2426 | No | ||
| 91 | RRAGC | 12093 | -0.424 | -0.2482 | No | ||
| 92 | HNRPF | 12193 | -0.439 | -0.2507 | No | ||
| 93 | RORC | 12213 | -0.443 | -0.2489 | No | ||
| 94 | FABP3 | 12305 | -0.453 | -0.2509 | No | ||
| 95 | LMX1A | 12485 | -0.481 | -0.2576 | No | ||
| 96 | CPT1A | 12538 | -0.489 | -0.2572 | No | ||
| 97 | ALX4 | 12602 | -0.498 | -0.2575 | No | ||
| 98 | FADS3 | 12800 | -0.529 | -0.2648 | No | ||
| 99 | RBBP4 | 12833 | -0.536 | -0.2631 | No | ||
| 100 | POU3F1 | 12907 | -0.550 | -0.2635 | No | ||
| 101 | GPD1 | 12982 | -0.561 | -0.2639 | No | ||
| 102 | HPCA | 13019 | -0.568 | -0.2622 | No | ||
| 103 | GTF2H1 | 13034 | -0.570 | -0.2593 | No | ||
| 104 | SLC16A1 | 13057 | -0.573 | -0.2568 | No | ||
| 105 | PAX6 | 13085 | -0.576 | -0.2546 | No | ||
| 106 | CSDA | 13145 | -0.585 | -0.2541 | No | ||
| 107 | COL2A1 | 13202 | -0.594 | -0.2533 | No | ||
| 108 | SORL1 | 13787 | -0.698 | -0.2805 | No | ||
| 109 | SC5DL | 13818 | -0.705 | -0.2776 | No | ||
| 110 | RPL22 | 13948 | -0.727 | -0.2799 | No | ||
| 111 | CBARA1 | 14162 | -0.766 | -0.2865 | No | ||
| 112 | WEE1 | 14185 | -0.770 | -0.2828 | No | ||
| 113 | NIT1 | 14218 | -0.776 | -0.2795 | No | ||
| 114 | HLX1 | 14236 | -0.781 | -0.2755 | No | ||
| 115 | POGK | 14248 | -0.783 | -0.2710 | No | ||
| 116 | SYT6 | 14907 | -0.914 | -0.3008 | No | ||
| 117 | FXYD6 | 15103 | -0.954 | -0.3053 | Yes | ||
| 118 | NRAS | 15105 | -0.954 | -0.2992 | Yes | ||
| 119 | UBE4B | 15107 | -0.954 | -0.2932 | Yes | ||
| 120 | HNRPA1 | 15361 | -1.006 | -0.3004 | Yes | ||
| 121 | ARHGAP12 | 15375 | -1.009 | -0.2947 | Yes | ||
| 122 | TGFB2 | 15506 | -1.035 | -0.2951 | Yes | ||
| 123 | TAGLN2 | 15518 | -1.039 | -0.2890 | Yes | ||
| 124 | CUL5 | 15601 | -1.056 | -0.2867 | Yes | ||
| 125 | EBNA1BP2 | 15716 | -1.080 | -0.2860 | Yes | ||
| 126 | TIMM10 | 15731 | -1.082 | -0.2798 | Yes | ||
| 127 | FOXD3 | 15840 | -1.109 | -0.2785 | Yes | ||
| 128 | FKBP11 | 15858 | -1.116 | -0.2723 | Yes | ||
| 129 | STMN1 | 15992 | -1.152 | -0.2721 | Yes | ||
| 130 | LRFN4 | 16059 | -1.167 | -0.2682 | Yes | ||
| 131 | LTBR | 16137 | -1.185 | -0.2648 | Yes | ||
| 132 | ATP6V0B | 16195 | -1.200 | -0.2602 | Yes | ||
| 133 | RAB3IL1 | 16222 | -1.208 | -0.2539 | Yes | ||
| 134 | UBXD3 | 16312 | -1.236 | -0.2508 | Yes | ||
| 135 | BAI2 | 16357 | -1.246 | -0.2452 | Yes | ||
| 136 | SLC38A2 | 16430 | -1.262 | -0.2410 | Yes | ||
| 137 | NKX2-3 | 16434 | -1.263 | -0.2330 | Yes | ||
| 138 | CBX5 | 16548 | -1.294 | -0.2309 | Yes | ||
| 139 | COMMD3 | 16815 | -1.377 | -0.2365 | Yes | ||
| 140 | CHD4 | 16895 | -1.401 | -0.2318 | Yes | ||
| 141 | ATF7IP | 16980 | -1.430 | -0.2272 | Yes | ||
| 142 | RHEBL1 | 17052 | -1.457 | -0.2217 | Yes | ||
| 143 | SCAMP3 | 17158 | -1.498 | -0.2178 | Yes | ||
| 144 | CTBP2 | 17366 | -1.590 | -0.2188 | Yes | ||
| 145 | AK2 | 17412 | -1.610 | -0.2109 | Yes | ||
| 146 | ARPC5 | 17481 | -1.634 | -0.2041 | Yes | ||
| 147 | H3F3A | 17616 | -1.701 | -0.2005 | Yes | ||
| 148 | TFB2M | 17674 | -1.729 | -0.1925 | Yes | ||
| 149 | CUGBP1 | 17737 | -1.767 | -0.1845 | Yes | ||
| 150 | EIF4B | 17836 | -1.829 | -0.1781 | Yes | ||
| 151 | STX6 | 17967 | -1.920 | -0.1728 | Yes | ||
| 152 | HHEX | 18004 | -1.960 | -0.1622 | Yes | ||
| 153 | ADAM12 | 18040 | -1.990 | -0.1514 | Yes | ||
| 154 | PABPC4 | 18152 | -2.106 | -0.1439 | Yes | ||
| 155 | NUDC | 18192 | -2.156 | -0.1322 | Yes | ||
| 156 | IVNS1ABP | 18228 | -2.201 | -0.1200 | Yes | ||
| 157 | SFXN2 | 18352 | -2.385 | -0.1114 | Yes | ||
| 158 | PFDN2 | 18360 | -2.392 | -0.0964 | Yes | ||
| 159 | FAF1 | 18444 | -2.593 | -0.0843 | Yes | ||
| 160 | EPC1 | 18540 | -2.994 | -0.0702 | Yes | ||
| 161 | ADK | 18597 | -3.603 | -0.0502 | Yes | ||
| 162 | CTSF | 18599 | -3.667 | -0.0267 | Yes | ||
| 163 | ALDH3B1 | 18614 | -4.308 | 0.0001 | Yes |