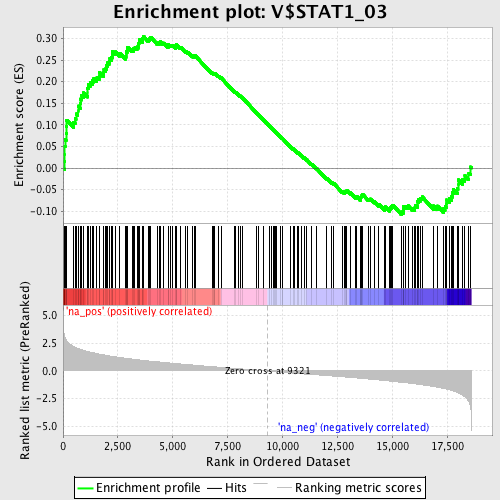
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$STAT1_03 |
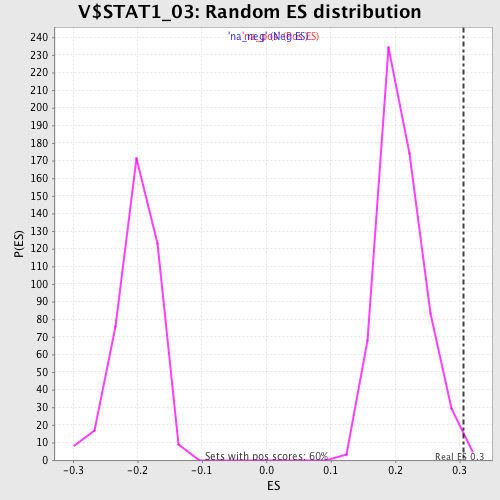
| Enrichment Score (ES) | 0.30565166 |
| Normalized Enrichment Score (NES) | 1.455068 |
| Nominal p-value | 0.008389262 |
| FDR q-value | 0.48050982 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HIVEP3 | 62 | 3.166 | 0.0149 | Yes | ||
| 2 | VPS41 | 71 | 3.107 | 0.0324 | Yes | ||
| 3 | PYY | 76 | 3.090 | 0.0500 | Yes | ||
| 4 | SLITRK3 | 93 | 3.012 | 0.0665 | Yes | ||
| 5 | IRF1 | 136 | 2.824 | 0.0805 | Yes | ||
| 6 | CCND1 | 152 | 2.793 | 0.0958 | Yes | ||
| 7 | EIF2B4 | 165 | 2.740 | 0.1110 | Yes | ||
| 8 | GRK5 | 483 | 2.207 | 0.1065 | Yes | ||
| 9 | GRIK1 | 548 | 2.144 | 0.1154 | Yes | ||
| 10 | PDE6D | 589 | 2.099 | 0.1254 | Yes | ||
| 11 | XLKD1 | 678 | 2.020 | 0.1322 | Yes | ||
| 12 | HOXB6 | 681 | 2.018 | 0.1438 | Yes | ||
| 13 | DDX25 | 780 | 1.951 | 0.1497 | Yes | ||
| 14 | ODF1 | 798 | 1.943 | 0.1600 | Yes | ||
| 15 | EPHA1 | 844 | 1.911 | 0.1686 | Yes | ||
| 16 | BDNF | 938 | 1.854 | 0.1742 | Yes | ||
| 17 | GNG4 | 1119 | 1.753 | 0.1746 | Yes | ||
| 18 | FBS1 | 1120 | 1.753 | 0.1847 | Yes | ||
| 19 | WWOX | 1149 | 1.739 | 0.1932 | Yes | ||
| 20 | ERG | 1231 | 1.700 | 0.1986 | Yes | ||
| 21 | PIK4CB | 1332 | 1.657 | 0.2028 | Yes | ||
| 22 | SALL1 | 1401 | 1.632 | 0.2085 | Yes | ||
| 23 | PPP1R9B | 1543 | 1.577 | 0.2099 | Yes | ||
| 24 | AFP | 1643 | 1.531 | 0.2134 | Yes | ||
| 25 | RHOG | 1661 | 1.524 | 0.2213 | Yes | ||
| 26 | ELOVL6 | 1829 | 1.462 | 0.2207 | Yes | ||
| 27 | NTRK3 | 1860 | 1.450 | 0.2274 | Yes | ||
| 28 | KLK9 | 1950 | 1.416 | 0.2308 | Yes | ||
| 29 | CBX3 | 1985 | 1.403 | 0.2370 | Yes | ||
| 30 | DOCK3 | 2010 | 1.393 | 0.2437 | Yes | ||
| 31 | EXTL3 | 2094 | 1.368 | 0.2471 | Yes | ||
| 32 | GRIA1 | 2132 | 1.353 | 0.2529 | Yes | ||
| 33 | BCLAF1 | 2203 | 1.327 | 0.2568 | Yes | ||
| 34 | RGS17 | 2238 | 1.317 | 0.2625 | Yes | ||
| 35 | IL23A | 2248 | 1.313 | 0.2696 | Yes | ||
| 36 | GALR3 | 2391 | 1.271 | 0.2693 | Yes | ||
| 37 | ARHGAP9 | 2581 | 1.219 | 0.2661 | Yes | ||
| 38 | GABRA1 | 2836 | 1.152 | 0.2589 | Yes | ||
| 39 | UBE2N | 2869 | 1.144 | 0.2638 | Yes | ||
| 40 | SNX17 | 2871 | 1.143 | 0.2703 | Yes | ||
| 41 | RELA | 2917 | 1.129 | 0.2744 | Yes | ||
| 42 | TRAF4 | 2933 | 1.126 | 0.2801 | Yes | ||
| 43 | ZAP70 | 3141 | 1.073 | 0.2751 | Yes | ||
| 44 | DDR1 | 3221 | 1.056 | 0.2769 | Yes | ||
| 45 | GSK3A | 3275 | 1.042 | 0.2800 | Yes | ||
| 46 | HSD17B4 | 3371 | 1.020 | 0.2807 | Yes | ||
| 47 | VAX1 | 3423 | 1.009 | 0.2838 | Yes | ||
| 48 | RAB8B | 3448 | 1.003 | 0.2883 | Yes | ||
| 49 | SLC30A3 | 3462 | 0.999 | 0.2933 | Yes | ||
| 50 | NUCB1 | 3494 | 0.992 | 0.2974 | Yes | ||
| 51 | PHF15 | 3621 | 0.962 | 0.2961 | Yes | ||
| 52 | USP2 | 3631 | 0.960 | 0.3012 | Yes | ||
| 53 | CLDN5 | 3651 | 0.957 | 0.3057 | Yes | ||
| 54 | PCQAP | 3883 | 0.905 | 0.2984 | No | ||
| 55 | RKHD3 | 3954 | 0.890 | 0.2997 | No | ||
| 56 | ITPKC | 3986 | 0.882 | 0.3031 | No | ||
| 57 | DTX2 | 4314 | 0.821 | 0.2901 | No | ||
| 58 | GRIN2B | 4378 | 0.811 | 0.2914 | No | ||
| 59 | DLX1 | 4421 | 0.802 | 0.2937 | No | ||
| 60 | PDE4D | 4578 | 0.772 | 0.2897 | No | ||
| 61 | GDF7 | 4787 | 0.732 | 0.2827 | No | ||
| 62 | ADCK4 | 4805 | 0.730 | 0.2860 | No | ||
| 63 | SFRS2 | 4907 | 0.712 | 0.2846 | No | ||
| 64 | EPHB2 | 4978 | 0.698 | 0.2848 | No | ||
| 65 | CHRM1 | 5138 | 0.669 | 0.2801 | No | ||
| 66 | RAPH1 | 5160 | 0.665 | 0.2828 | No | ||
| 67 | CCNI | 5174 | 0.662 | 0.2859 | No | ||
| 68 | NLK | 5345 | 0.634 | 0.2803 | No | ||
| 69 | NKX2-8 | 5583 | 0.591 | 0.2709 | No | ||
| 70 | POLR1B | 5688 | 0.574 | 0.2686 | No | ||
| 71 | COX8A | 5899 | 0.541 | 0.2603 | No | ||
| 72 | EGF | 5982 | 0.525 | 0.2589 | No | ||
| 73 | DNAJA2 | 6011 | 0.521 | 0.2604 | No | ||
| 74 | HOXC4 | 6793 | 0.391 | 0.2203 | No | ||
| 75 | ERF | 6871 | 0.378 | 0.2183 | No | ||
| 76 | PLCB2 | 6892 | 0.375 | 0.2194 | No | ||
| 77 | NHLH2 | 6920 | 0.371 | 0.2201 | No | ||
| 78 | EN1 | 7089 | 0.344 | 0.2130 | No | ||
| 79 | IGFALS | 7196 | 0.326 | 0.2091 | No | ||
| 80 | HOXB2 | 7822 | 0.233 | 0.1766 | No | ||
| 81 | PCF11 | 7863 | 0.227 | 0.1757 | No | ||
| 82 | DHH | 7994 | 0.206 | 0.1699 | No | ||
| 83 | FBXW11 | 8067 | 0.194 | 0.1671 | No | ||
| 84 | GALK2 | 8154 | 0.182 | 0.1635 | No | ||
| 85 | TLK2 | 8825 | 0.074 | 0.1276 | No | ||
| 86 | ZNRF1 | 8889 | 0.063 | 0.1245 | No | ||
| 87 | EIF4G1 | 9140 | 0.028 | 0.1112 | No | ||
| 88 | ITM2C | 9402 | -0.013 | 0.0971 | No | ||
| 89 | STX16 | 9477 | -0.024 | 0.0932 | No | ||
| 90 | MRPL54 | 9569 | -0.037 | 0.0885 | No | ||
| 91 | GATA3 | 9615 | -0.046 | 0.0863 | No | ||
| 92 | SEC14L2 | 9676 | -0.057 | 0.0834 | No | ||
| 93 | PSMC3 | 9742 | -0.070 | 0.0803 | No | ||
| 94 | MYH10 | 9894 | -0.098 | 0.0727 | No | ||
| 95 | PABPN1 | 9992 | -0.113 | 0.0681 | No | ||
| 96 | MRPL34 | 9996 | -0.114 | 0.0686 | No | ||
| 97 | FOXA3 | 10377 | -0.169 | 0.0489 | No | ||
| 98 | FEM1A | 10477 | -0.182 | 0.0446 | No | ||
| 99 | PCDHGB5 | 10560 | -0.196 | 0.0413 | No | ||
| 100 | COL16A1 | 10691 | -0.216 | 0.0355 | No | ||
| 101 | SOX4 | 10722 | -0.220 | 0.0352 | No | ||
| 102 | PIK3CD | 10858 | -0.244 | 0.0293 | No | ||
| 103 | MAG | 10990 | -0.267 | 0.0237 | No | ||
| 104 | CS | 11102 | -0.283 | 0.0193 | No | ||
| 105 | HSPD1 | 11322 | -0.316 | 0.0093 | No | ||
| 106 | SMPD3 | 11540 | -0.343 | -0.0005 | No | ||
| 107 | RDH11 | 12008 | -0.411 | -0.0235 | No | ||
| 108 | CNTN6 | 12245 | -0.447 | -0.0337 | No | ||
| 109 | C4BPA | 12322 | -0.456 | -0.0351 | No | ||
| 110 | SLC9A1 | 12733 | -0.519 | -0.0544 | No | ||
| 111 | BRCA1 | 12837 | -0.537 | -0.0569 | No | ||
| 112 | SPTLC2 | 12856 | -0.541 | -0.0547 | No | ||
| 113 | PPP2CB | 12896 | -0.548 | -0.0537 | No | ||
| 114 | TTC1 | 12934 | -0.554 | -0.0525 | No | ||
| 115 | PAX6 | 13085 | -0.576 | -0.0573 | No | ||
| 116 | PRKACA | 13345 | -0.621 | -0.0677 | No | ||
| 117 | RPL28 | 13384 | -0.627 | -0.0662 | No | ||
| 118 | APBA3 | 13542 | -0.657 | -0.0709 | No | ||
| 119 | ADAM22 | 13572 | -0.661 | -0.0687 | No | ||
| 120 | TCERG1 | 13581 | -0.663 | -0.0653 | No | ||
| 121 | SMARCA1 | 13616 | -0.667 | -0.0633 | No | ||
| 122 | NXPH4 | 13646 | -0.673 | -0.0610 | No | ||
| 123 | NCOA3 | 13927 | -0.723 | -0.0720 | No | ||
| 124 | GFI1 | 14001 | -0.734 | -0.0717 | No | ||
| 125 | BET1 | 14177 | -0.769 | -0.0767 | No | ||
| 126 | CRK | 14395 | -0.808 | -0.0838 | No | ||
| 127 | STC1 | 14660 | -0.861 | -0.0932 | No | ||
| 128 | ZNF385 | 14693 | -0.868 | -0.0899 | No | ||
| 129 | MAP3K8 | 14878 | -0.909 | -0.0946 | No | ||
| 130 | SYT6 | 14907 | -0.914 | -0.0909 | No | ||
| 131 | SEMA4B | 14959 | -0.926 | -0.0883 | No | ||
| 132 | SLAMF1 | 15015 | -0.938 | -0.0859 | No | ||
| 133 | TBX5 | 15418 | -1.017 | -0.1018 | No | ||
| 134 | LRP5 | 15497 | -1.034 | -0.1000 | No | ||
| 135 | GNB3 | 15501 | -1.034 | -0.0942 | No | ||
| 136 | JUB | 15520 | -1.039 | -0.0892 | No | ||
| 137 | RPS27 | 15626 | -1.060 | -0.0888 | No | ||
| 138 | DLL1 | 15720 | -1.080 | -0.0876 | No | ||
| 139 | GADD45B | 15938 | -1.139 | -0.0928 | No | ||
| 140 | UBL3 | 16036 | -1.163 | -0.0913 | No | ||
| 141 | LRFN4 | 16059 | -1.167 | -0.0858 | No | ||
| 142 | SIN3A | 16152 | -1.191 | -0.0839 | No | ||
| 143 | SNX9 | 16154 | -1.191 | -0.0771 | No | ||
| 144 | CDK6 | 16209 | -1.205 | -0.0731 | No | ||
| 145 | HNRPA2B1 | 16276 | -1.227 | -0.0696 | No | ||
| 146 | E2F3 | 16358 | -1.247 | -0.0668 | No | ||
| 147 | CHD4 | 16895 | -1.401 | -0.0877 | No | ||
| 148 | HSPE1 | 17055 | -1.458 | -0.0879 | No | ||
| 149 | ARPC1B | 17344 | -1.580 | -0.0944 | No | ||
| 150 | NRG1 | 17407 | -1.609 | -0.0885 | No | ||
| 151 | SKP2 | 17483 | -1.636 | -0.0831 | No | ||
| 152 | GSPT1 | 17489 | -1.638 | -0.0740 | No | ||
| 153 | DMD | 17599 | -1.693 | -0.0701 | No | ||
| 154 | DDX47 | 17714 | -1.747 | -0.0662 | No | ||
| 155 | CD9 | 17736 | -1.766 | -0.0572 | No | ||
| 156 | CCR4 | 17789 | -1.799 | -0.0496 | No | ||
| 157 | STX6 | 17967 | -1.920 | -0.0481 | No | ||
| 158 | HDAC9 | 17999 | -1.952 | -0.0385 | No | ||
| 159 | HHEX | 18004 | -1.960 | -0.0275 | No | ||
| 160 | H1F0 | 18207 | -2.177 | -0.0258 | No | ||
| 161 | VCAM1 | 18314 | -2.331 | -0.0181 | No | ||
| 162 | CENTA2 | 18492 | -2.777 | -0.0117 | No | ||
| 163 | CSNK1A1 | 18570 | -3.187 | 0.0025 | No |