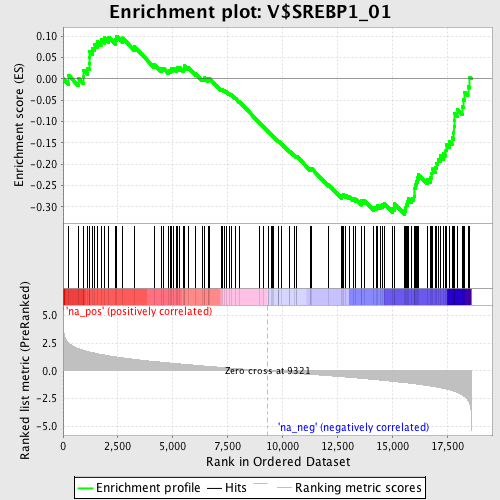
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$SREBP1_01 |
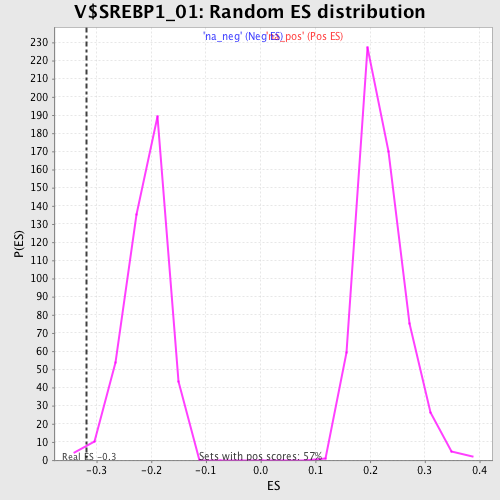
| Enrichment Score (ES) | -0.3176144 |
| Normalized Enrichment Score (NES) | -1.5204929 |
| Nominal p-value | 0.009195402 |
| FDR q-value | 0.21542308 |
| FWER p-Value | 0.96 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP6V0D1 | 258 | 2.503 | 0.0076 | No | ||
| 2 | ABR | 701 | 2.003 | 0.0009 | No | ||
| 3 | SLC35A5 | 915 | 1.865 | 0.0055 | No | ||
| 4 | BDNF | 938 | 1.854 | 0.0202 | No | ||
| 5 | ANAPC13 | 1128 | 1.746 | 0.0251 | No | ||
| 6 | ZFYVE26 | 1191 | 1.716 | 0.0365 | No | ||
| 7 | HNRPH2 | 1202 | 1.712 | 0.0507 | No | ||
| 8 | SOX10 | 1210 | 1.709 | 0.0650 | No | ||
| 9 | LCP1 | 1350 | 1.649 | 0.0717 | No | ||
| 10 | DIRC2 | 1443 | 1.615 | 0.0806 | No | ||
| 11 | HIVEP1 | 1575 | 1.561 | 0.0870 | No | ||
| 12 | DUSP8 | 1730 | 1.500 | 0.0915 | No | ||
| 13 | IRS4 | 1877 | 1.444 | 0.0961 | No | ||
| 14 | U2AF2 | 2078 | 1.374 | 0.0971 | No | ||
| 15 | HSPBAP1 | 2382 | 1.274 | 0.0917 | No | ||
| 16 | GLA | 2431 | 1.261 | 0.0999 | No | ||
| 17 | SDF2 | 2702 | 1.191 | 0.0956 | No | ||
| 18 | RAD9A | 3250 | 1.048 | 0.0750 | No | ||
| 19 | CCAR1 | 4166 | 0.851 | 0.0329 | No | ||
| 20 | ATF4 | 4469 | 0.794 | 0.0234 | No | ||
| 21 | SEPT3 | 4559 | 0.776 | 0.0253 | No | ||
| 22 | MTCH2 | 4803 | 0.730 | 0.0184 | No | ||
| 23 | HOXB7 | 4879 | 0.717 | 0.0205 | No | ||
| 24 | KIAA1715 | 4929 | 0.709 | 0.0240 | No | ||
| 25 | RNF146 | 5037 | 0.689 | 0.0241 | No | ||
| 26 | OTOP2 | 5170 | 0.663 | 0.0227 | No | ||
| 27 | SNX16 | 5217 | 0.655 | 0.0258 | No | ||
| 28 | AARS | 5324 | 0.638 | 0.0256 | No | ||
| 29 | DAZAP2 | 5469 | 0.609 | 0.0231 | No | ||
| 30 | RAB22A | 5517 | 0.601 | 0.0257 | No | ||
| 31 | RGL1 | 5528 | 0.599 | 0.0303 | No | ||
| 32 | MRPL27 | 5692 | 0.574 | 0.0264 | No | ||
| 33 | BLOC1S1 | 6035 | 0.516 | 0.0124 | No | ||
| 34 | DLX2 | 6352 | 0.463 | -0.0007 | No | ||
| 35 | USH1G | 6431 | 0.449 | -0.0011 | No | ||
| 36 | RAB3A | 6444 | 0.447 | 0.0021 | No | ||
| 37 | RRM2B | 6604 | 0.422 | -0.0028 | No | ||
| 38 | MYST2 | 6610 | 0.421 | 0.0005 | No | ||
| 39 | POLR3C | 6682 | 0.410 | 0.0002 | No | ||
| 40 | SLC36A1 | 7211 | 0.323 | -0.0256 | No | ||
| 41 | ATP7A | 7241 | 0.319 | -0.0244 | No | ||
| 42 | UBE2B | 7349 | 0.300 | -0.0276 | No | ||
| 43 | SNX2 | 7427 | 0.288 | -0.0293 | No | ||
| 44 | SUPT16H | 7584 | 0.265 | -0.0354 | No | ||
| 45 | DVL2 | 7665 | 0.254 | -0.0376 | No | ||
| 46 | HOXA7 | 7839 | 0.230 | -0.0449 | No | ||
| 47 | FEN1 | 8037 | 0.197 | -0.0539 | No | ||
| 48 | TRPM7 | 8944 | 0.056 | -0.1024 | No | ||
| 49 | EIF4G1 | 9140 | 0.028 | -0.1127 | No | ||
| 50 | HOXB5 | 9351 | -0.006 | -0.1240 | No | ||
| 51 | ATP6V1A | 9366 | -0.008 | -0.1247 | No | ||
| 52 | SNAI1 | 9498 | -0.027 | -0.1315 | No | ||
| 53 | QTRT1 | 9536 | -0.032 | -0.1333 | No | ||
| 54 | CHD2 | 9588 | -0.042 | -0.1357 | No | ||
| 55 | PIAS4 | 9799 | -0.081 | -0.1463 | No | ||
| 56 | TOP3A | 9808 | -0.082 | -0.1460 | No | ||
| 57 | GNB2 | 9823 | -0.085 | -0.1461 | No | ||
| 58 | XPO1 | 9942 | -0.106 | -0.1515 | No | ||
| 59 | PIP5K1A | 10312 | -0.158 | -0.1701 | No | ||
| 60 | PICALM | 10336 | -0.162 | -0.1700 | No | ||
| 61 | HOXC5 | 10534 | -0.191 | -0.1790 | No | ||
| 62 | HOXA2 | 10623 | -0.206 | -0.1820 | No | ||
| 63 | NGFRAP1 | 10651 | -0.209 | -0.1816 | No | ||
| 64 | SIRT1 | 11267 | -0.307 | -0.2122 | No | ||
| 65 | HTATIP | 11272 | -0.308 | -0.2098 | No | ||
| 66 | MCM2 | 11323 | -0.316 | -0.2098 | No | ||
| 67 | RRAGC | 12093 | -0.424 | -0.2477 | No | ||
| 68 | TOPORS | 12707 | -0.517 | -0.2764 | No | ||
| 69 | PDCD6IP | 12711 | -0.517 | -0.2721 | No | ||
| 70 | HOXA11 | 12763 | -0.524 | -0.2704 | No | ||
| 71 | CAMK2D | 12888 | -0.547 | -0.2724 | No | ||
| 72 | GTF2H1 | 13034 | -0.570 | -0.2753 | No | ||
| 73 | BAX | 13218 | -0.596 | -0.2801 | No | ||
| 74 | ASNA1 | 13341 | -0.620 | -0.2813 | No | ||
| 75 | AKAP12 | 13605 | -0.666 | -0.2898 | No | ||
| 76 | SNRPA | 13614 | -0.667 | -0.2845 | No | ||
| 77 | CSRP2BP | 13742 | -0.691 | -0.2854 | No | ||
| 78 | CBARA1 | 14162 | -0.766 | -0.3015 | No | ||
| 79 | SMCR8 | 14265 | -0.786 | -0.3002 | No | ||
| 80 | CD164 | 14336 | -0.800 | -0.2971 | No | ||
| 81 | HPS3 | 14477 | -0.825 | -0.2976 | No | ||
| 82 | GGN | 14555 | -0.841 | -0.2945 | No | ||
| 83 | FMR1 | 14625 | -0.854 | -0.2909 | No | ||
| 84 | SUPT6H | 15024 | -0.939 | -0.3043 | No | ||
| 85 | IGF2R | 15091 | -0.952 | -0.2997 | No | ||
| 86 | UBE4B | 15107 | -0.954 | -0.2923 | No | ||
| 87 | ALG1 | 15576 | -1.051 | -0.3086 | Yes | ||
| 88 | CUL5 | 15601 | -1.056 | -0.3008 | Yes | ||
| 89 | DSCAM | 15629 | -1.061 | -0.2931 | Yes | ||
| 90 | ADCY2 | 15710 | -1.079 | -0.2881 | Yes | ||
| 91 | ARMCX3 | 15735 | -1.083 | -0.2801 | Yes | ||
| 92 | ASPSCR1 | 15900 | -1.128 | -0.2793 | Yes | ||
| 93 | STMN1 | 15992 | -1.152 | -0.2743 | Yes | ||
| 94 | SET | 16028 | -1.161 | -0.2662 | Yes | ||
| 95 | APEX1 | 16029 | -1.161 | -0.2562 | Yes | ||
| 96 | POLDIP2 | 16052 | -1.165 | -0.2473 | Yes | ||
| 97 | GRN | 16091 | -1.175 | -0.2393 | Yes | ||
| 98 | GSK3B | 16146 | -1.190 | -0.2320 | Yes | ||
| 99 | PRKCE | 16205 | -1.204 | -0.2247 | Yes | ||
| 100 | OSGEP | 16596 | -1.308 | -0.2346 | Yes | ||
| 101 | ZDHHC13 | 16739 | -1.355 | -0.2306 | Yes | ||
| 102 | CLN3 | 16801 | -1.371 | -0.2221 | Yes | ||
| 103 | COMMD3 | 16815 | -1.377 | -0.2109 | Yes | ||
| 104 | ATF7IP | 16980 | -1.430 | -0.2075 | Yes | ||
| 105 | ALDH6A1 | 17024 | -1.445 | -0.1974 | Yes | ||
| 106 | ZIC3 | 17086 | -1.469 | -0.1880 | Yes | ||
| 107 | TCEAL1 | 17185 | -1.509 | -0.1803 | Yes | ||
| 108 | ACCN1 | 17342 | -1.578 | -0.1752 | Yes | ||
| 109 | GPNMB | 17447 | -1.622 | -0.1668 | Yes | ||
| 110 | ARPC5 | 17481 | -1.634 | -0.1546 | Yes | ||
| 111 | LAMP1 | 17606 | -1.697 | -0.1466 | Yes | ||
| 112 | BLNK | 17729 | -1.762 | -0.1381 | Yes | ||
| 113 | DCTN4 | 17805 | -1.806 | -0.1266 | Yes | ||
| 114 | SOCS2 | 17815 | -1.816 | -0.1114 | Yes | ||
| 115 | EIF4B | 17836 | -1.829 | -0.0968 | Yes | ||
| 116 | HK2 | 17843 | -1.836 | -0.0813 | Yes | ||
| 117 | STX6 | 17967 | -1.920 | -0.0714 | Yes | ||
| 118 | TNRC15 | 18202 | -2.165 | -0.0654 | Yes | ||
| 119 | CANX | 18244 | -2.219 | -0.0485 | Yes | ||
| 120 | KLF12 | 18302 | -2.309 | -0.0317 | Yes | ||
| 121 | NR1D1 | 18454 | -2.653 | -0.0171 | Yes | ||
| 122 | EPC1 | 18540 | -2.994 | 0.0041 | Yes |