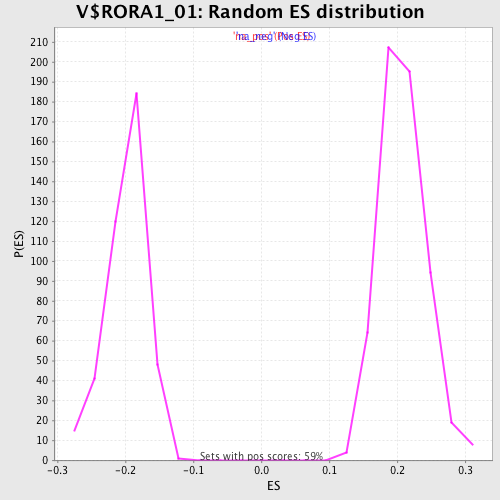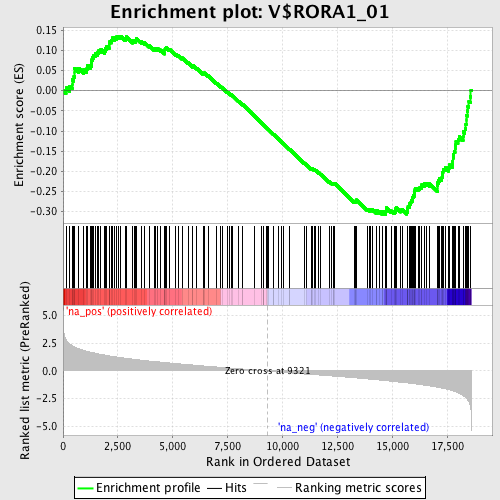
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$RORA1_01 |
| Enrichment Score (ES) | -0.30689064 |
| Normalized Enrichment Score (NES) | -1.5446631 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.21178819 |
| FWER p-Value | 0.917 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MPP3 | 133 | 2.834 | 0.0072 | No | ||
| 2 | ATP1B4 | 294 | 2.455 | 0.0110 | No | ||
| 3 | ASPH | 414 | 2.287 | 0.0162 | No | ||
| 4 | JMJD1C | 418 | 2.283 | 0.0276 | No | ||
| 5 | KCNAB3 | 493 | 2.193 | 0.0347 | No | ||
| 6 | VAMP1 | 521 | 2.168 | 0.0443 | No | ||
| 7 | ERN1 | 523 | 2.165 | 0.0552 | No | ||
| 8 | CHRDL1 | 706 | 2.001 | 0.0555 | No | ||
| 9 | GRIK3 | 945 | 1.849 | 0.0520 | No | ||
| 10 | FBXW7 | 1077 | 1.772 | 0.0539 | No | ||
| 11 | SORCS1 | 1095 | 1.762 | 0.0620 | No | ||
| 12 | ENO3 | 1236 | 1.698 | 0.0630 | No | ||
| 13 | SLC8A3 | 1287 | 1.678 | 0.0688 | No | ||
| 14 | RASAL1 | 1307 | 1.669 | 0.0763 | No | ||
| 15 | PPP2R5D | 1346 | 1.650 | 0.0826 | No | ||
| 16 | EPHA7 | 1396 | 1.634 | 0.0882 | No | ||
| 17 | RGS6 | 1474 | 1.602 | 0.0922 | No | ||
| 18 | UBADC1 | 1581 | 1.558 | 0.0944 | No | ||
| 19 | SLC18A2 | 1630 | 1.537 | 0.0996 | No | ||
| 20 | PITPNM1 | 1720 | 1.504 | 0.1024 | No | ||
| 21 | SUCLA2 | 1907 | 1.431 | 0.0996 | No | ||
| 22 | ATP1B2 | 1940 | 1.419 | 0.1051 | No | ||
| 23 | DTNA | 1991 | 1.401 | 0.1095 | No | ||
| 24 | STK38 | 2098 | 1.367 | 0.1107 | No | ||
| 25 | ATP8B2 | 2099 | 1.366 | 0.1176 | No | ||
| 26 | GRIA1 | 2132 | 1.353 | 0.1228 | No | ||
| 27 | SCOTIN | 2217 | 1.325 | 0.1249 | No | ||
| 28 | ANGPT1 | 2230 | 1.321 | 0.1310 | No | ||
| 29 | ELAVL4 | 2348 | 1.285 | 0.1312 | No | ||
| 30 | IL17F | 2428 | 1.261 | 0.1333 | No | ||
| 31 | JARID2 | 2508 | 1.238 | 0.1353 | No | ||
| 32 | PBXIP1 | 2635 | 1.206 | 0.1346 | No | ||
| 33 | VAMP2 | 2835 | 1.152 | 0.1297 | No | ||
| 34 | HOXB3 | 2870 | 1.143 | 0.1336 | No | ||
| 35 | HCN1 | 3158 | 1.068 | 0.1235 | No | ||
| 36 | ACSL4 | 3255 | 1.046 | 0.1236 | No | ||
| 37 | CAMK2B | 3321 | 1.030 | 0.1253 | No | ||
| 38 | CNTFR | 3347 | 1.024 | 0.1292 | No | ||
| 39 | KHDRBS2 | 3574 | 0.972 | 0.1218 | No | ||
| 40 | TTL | 3700 | 0.945 | 0.1199 | No | ||
| 41 | PHF5A | 3925 | 0.894 | 0.1123 | No | ||
| 42 | SLC22A6 | 4159 | 0.852 | 0.1040 | No | ||
| 43 | RNF12 | 4211 | 0.842 | 0.1055 | No | ||
| 44 | AP3D1 | 4310 | 0.823 | 0.1043 | No | ||
| 45 | ZDHHC21 | 4447 | 0.798 | 0.1010 | No | ||
| 46 | B3GALT2 | 4635 | 0.760 | 0.0947 | No | ||
| 47 | HOXA3 | 4639 | 0.759 | 0.0984 | No | ||
| 48 | TPM3 | 4640 | 0.759 | 0.1023 | No | ||
| 49 | PKP3 | 4660 | 0.756 | 0.1051 | No | ||
| 50 | TIMM9 | 4696 | 0.749 | 0.1070 | No | ||
| 51 | NR2F2 | 4837 | 0.726 | 0.1031 | No | ||
| 52 | GPR23 | 5143 | 0.668 | 0.0900 | No | ||
| 53 | PSIP1 | 5249 | 0.649 | 0.0876 | No | ||
| 54 | RCOR1 | 5423 | 0.618 | 0.0813 | No | ||
| 55 | MRPL27 | 5692 | 0.574 | 0.0697 | No | ||
| 56 | SOX15 | 5905 | 0.540 | 0.0609 | No | ||
| 57 | DBP | 5911 | 0.539 | 0.0634 | No | ||
| 58 | BZW2 | 6094 | 0.504 | 0.0561 | No | ||
| 59 | HIST1H4C | 6400 | 0.454 | 0.0419 | No | ||
| 60 | ALS2CR2 | 6409 | 0.453 | 0.0437 | No | ||
| 61 | RAB3A | 6444 | 0.447 | 0.0442 | No | ||
| 62 | CSAD | 6607 | 0.422 | 0.0375 | No | ||
| 63 | CXXC5 | 6987 | 0.360 | 0.0188 | No | ||
| 64 | EFNA3 | 7178 | 0.330 | 0.0102 | No | ||
| 65 | ADCYAP1 | 7262 | 0.315 | 0.0073 | No | ||
| 66 | PACSIN1 | 7491 | 0.279 | -0.0037 | No | ||
| 67 | GEMIN7 | 7602 | 0.261 | -0.0083 | No | ||
| 68 | SAG | 7658 | 0.255 | -0.0100 | No | ||
| 69 | RG9MTD2 | 7719 | 0.244 | -0.0120 | No | ||
| 70 | JMJD1B | 7996 | 0.205 | -0.0260 | No | ||
| 71 | AMY2A | 8160 | 0.182 | -0.0339 | No | ||
| 72 | VASP | 8195 | 0.177 | -0.0348 | No | ||
| 73 | ARNTL | 8710 | 0.091 | -0.0622 | No | ||
| 74 | FLI1 | 9059 | 0.040 | -0.0809 | No | ||
| 75 | CACNA1A | 9120 | 0.031 | -0.0840 | No | ||
| 76 | NDRG2 | 9123 | 0.030 | -0.0840 | No | ||
| 77 | NOL4 | 9136 | 0.029 | -0.0845 | No | ||
| 78 | BCL9 | 9270 | 0.008 | -0.0916 | No | ||
| 79 | MLLT6 | 9329 | -0.002 | -0.0948 | No | ||
| 80 | ATP6V1A | 9366 | -0.008 | -0.0967 | No | ||
| 81 | IL22 | 9583 | -0.040 | -0.1082 | No | ||
| 82 | CHD2 | 9588 | -0.042 | -0.1082 | No | ||
| 83 | IL7 | 9800 | -0.081 | -0.1192 | No | ||
| 84 | NPAS2 | 9933 | -0.104 | -0.1259 | No | ||
| 85 | ARX | 10050 | -0.122 | -0.1315 | No | ||
| 86 | ESRRB | 10332 | -0.161 | -0.1460 | No | ||
| 87 | CYP39A1 | 10333 | -0.161 | -0.1451 | No | ||
| 88 | FGF9 | 10987 | -0.266 | -0.1792 | No | ||
| 89 | ABHD3 | 11092 | -0.281 | -0.1834 | No | ||
| 90 | PCDH21 | 11312 | -0.314 | -0.1937 | No | ||
| 91 | HSPD1 | 11322 | -0.316 | -0.1926 | No | ||
| 92 | MCC | 11366 | -0.322 | -0.1933 | No | ||
| 93 | LCN2 | 11441 | -0.328 | -0.1956 | No | ||
| 94 | PPP2CA | 11493 | -0.336 | -0.1967 | No | ||
| 95 | GRM1 | 11644 | -0.356 | -0.2030 | No | ||
| 96 | GALT | 11749 | -0.372 | -0.2068 | No | ||
| 97 | ATP5G1 | 12127 | -0.431 | -0.2250 | No | ||
| 98 | CNTN6 | 12245 | -0.447 | -0.2291 | No | ||
| 99 | MYL3 | 12331 | -0.457 | -0.2314 | No | ||
| 100 | ITGAX | 12338 | -0.457 | -0.2294 | No | ||
| 101 | PCDH18 | 12378 | -0.464 | -0.2291 | No | ||
| 102 | SHH | 13272 | -0.607 | -0.2745 | No | ||
| 103 | GABARAPL1 | 13314 | -0.615 | -0.2736 | No | ||
| 104 | MEF2C | 13348 | -0.622 | -0.2722 | No | ||
| 105 | NOG | 13381 | -0.627 | -0.2708 | No | ||
| 106 | SCMH1 | 13885 | -0.716 | -0.2944 | No | ||
| 107 | ATP6AP2 | 13966 | -0.730 | -0.2950 | No | ||
| 108 | MATK | 14004 | -0.734 | -0.2933 | No | ||
| 109 | YWHAQ | 14103 | -0.753 | -0.2948 | No | ||
| 110 | A2BP1 | 14278 | -0.789 | -0.3002 | No | ||
| 111 | NFATC3 | 14283 | -0.790 | -0.2964 | No | ||
| 112 | SPOCK2 | 14418 | -0.812 | -0.2996 | No | ||
| 113 | ACOXL | 14554 | -0.840 | -0.3026 | Yes | ||
| 114 | TRAP1 | 14561 | -0.842 | -0.2987 | Yes | ||
| 115 | MRVI1 | 14707 | -0.872 | -0.3021 | Yes | ||
| 116 | USP8 | 14716 | -0.874 | -0.2981 | Yes | ||
| 117 | SNTG1 | 14727 | -0.877 | -0.2942 | Yes | ||
| 118 | NFYA | 14738 | -0.877 | -0.2903 | Yes | ||
| 119 | ACO2 | 14971 | -0.928 | -0.2981 | Yes | ||
| 120 | MYB | 15099 | -0.953 | -0.3002 | Yes | ||
| 121 | NELL2 | 15143 | -0.960 | -0.2976 | Yes | ||
| 122 | LARGE | 15147 | -0.960 | -0.2929 | Yes | ||
| 123 | CAMKK2 | 15172 | -0.966 | -0.2893 | Yes | ||
| 124 | STAC2 | 15385 | -1.011 | -0.2957 | Yes | ||
| 125 | ESPN | 15449 | -1.024 | -0.2939 | Yes | ||
| 126 | AFG3L2 | 15673 | -1.069 | -0.3005 | Yes | ||
| 127 | SMPX | 15701 | -1.075 | -0.2965 | Yes | ||
| 128 | MPP6 | 15703 | -1.075 | -0.2911 | Yes | ||
| 129 | NRL | 15718 | -1.080 | -0.2864 | Yes | ||
| 130 | CALCA | 15796 | -1.098 | -0.2850 | Yes | ||
| 131 | AOC2 | 15798 | -1.098 | -0.2795 | Yes | ||
| 132 | PIM1 | 15850 | -1.112 | -0.2766 | Yes | ||
| 133 | TCF7 | 15861 | -1.117 | -0.2715 | Yes | ||
| 134 | SATB1 | 15911 | -1.132 | -0.2684 | Yes | ||
| 135 | KIF5A | 15931 | -1.138 | -0.2636 | Yes | ||
| 136 | GTF2IRD1 | 15967 | -1.147 | -0.2597 | Yes | ||
| 137 | CX3CL1 | 16001 | -1.156 | -0.2556 | Yes | ||
| 138 | SLC25A27 | 16011 | -1.157 | -0.2502 | Yes | ||
| 139 | UBL3 | 16036 | -1.163 | -0.2456 | Yes | ||
| 140 | LDB2 | 16068 | -1.170 | -0.2413 | Yes | ||
| 141 | IL17RE | 16185 | -1.199 | -0.2415 | Yes | ||
| 142 | RBMS1 | 16261 | -1.222 | -0.2394 | Yes | ||
| 143 | ZRANB1 | 16323 | -1.237 | -0.2364 | Yes | ||
| 144 | NEO1 | 16353 | -1.246 | -0.2316 | Yes | ||
| 145 | ARF6 | 16458 | -1.269 | -0.2308 | Yes | ||
| 146 | RYR1 | 16545 | -1.293 | -0.2289 | Yes | ||
| 147 | CNTN2 | 16708 | -1.346 | -0.2309 | Yes | ||
| 148 | ATP5J2 | 17054 | -1.458 | -0.2422 | Yes | ||
| 149 | HSPE1 | 17055 | -1.458 | -0.2348 | Yes | ||
| 150 | SLC4A7 | 17075 | -1.466 | -0.2283 | Yes | ||
| 151 | CTH | 17101 | -1.476 | -0.2222 | Yes | ||
| 152 | SPAG9 | 17160 | -1.499 | -0.2177 | Yes | ||
| 153 | CACNB2 | 17265 | -1.546 | -0.2155 | Yes | ||
| 154 | UHRF2 | 17281 | -1.552 | -0.2084 | Yes | ||
| 155 | DACT1 | 17304 | -1.562 | -0.2017 | Yes | ||
| 156 | PLA2G5 | 17316 | -1.569 | -0.1943 | Yes | ||
| 157 | ONECUT2 | 17405 | -1.608 | -0.1909 | Yes | ||
| 158 | BTBD3 | 17562 | -1.674 | -0.1909 | Yes | ||
| 159 | ATPAF2 | 17592 | -1.689 | -0.1839 | Yes | ||
| 160 | MSI2 | 17734 | -1.764 | -0.1825 | Yes | ||
| 161 | SEMA3F | 17759 | -1.780 | -0.1748 | Yes | ||
| 162 | ABAT | 17770 | -1.785 | -0.1663 | Yes | ||
| 163 | EXTL2 | 17786 | -1.798 | -0.1579 | Yes | ||
| 164 | SOCS2 | 17815 | -1.816 | -0.1502 | Yes | ||
| 165 | MYLK | 17863 | -1.848 | -0.1434 | Yes | ||
| 166 | NTF3 | 17872 | -1.854 | -0.1344 | Yes | ||
| 167 | YTHDF2 | 17904 | -1.878 | -0.1265 | Yes | ||
| 168 | HDAC9 | 17999 | -1.952 | -0.1217 | Yes | ||
| 169 | RASA4 | 18047 | -2.001 | -0.1141 | Yes | ||
| 170 | IVNS1ABP | 18228 | -2.201 | -0.1127 | Yes | ||
| 171 | DTNB | 18235 | -2.209 | -0.1018 | Yes | ||
| 172 | IBRDC2 | 18317 | -2.335 | -0.0943 | Yes | ||
| 173 | NAV1 | 18355 | -2.387 | -0.0842 | Yes | ||
| 174 | SLA | 18369 | -2.411 | -0.0726 | Yes | ||
| 175 | VNN3 | 18371 | -2.413 | -0.0604 | Yes | ||
| 176 | CNTF | 18418 | -2.516 | -0.0501 | Yes | ||
| 177 | ZFP36L1 | 18447 | -2.620 | -0.0383 | Yes | ||
| 178 | CHST1 | 18495 | -2.787 | -0.0267 | Yes | ||
| 179 | SOCS4 | 18562 | -3.083 | -0.0146 | Yes | ||
| 180 | CREB1 | 18587 | -3.440 | 0.0016 | Yes |