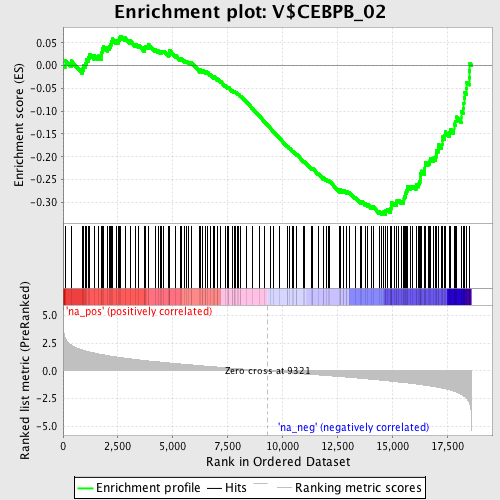
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$CEBPB_02 |
| Enrichment Score (ES) | -0.32654586 |
| Normalized Enrichment Score (NES) | -1.6504794 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.55830914 |
| FWER p-Value | 0.603 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAB3IP | 89 | 3.029 | 0.0121 | No | ||
| 2 | PRLR | 366 | 2.344 | 0.0102 | No | ||
| 3 | DDR2 | 885 | 1.886 | -0.0074 | No | ||
| 4 | BDNF | 938 | 1.854 | 0.0002 | No | ||
| 5 | TAC1 | 1033 | 1.798 | 0.0051 | No | ||
| 6 | FBXW7 | 1077 | 1.772 | 0.0127 | No | ||
| 7 | PKNOX2 | 1147 | 1.739 | 0.0187 | No | ||
| 8 | SOX10 | 1210 | 1.709 | 0.0248 | No | ||
| 9 | PITX2 | 1413 | 1.627 | 0.0230 | No | ||
| 10 | TRPM1 | 1607 | 1.548 | 0.0212 | No | ||
| 11 | COL4A4 | 1748 | 1.494 | 0.0219 | No | ||
| 12 | PLA2G2E | 1763 | 1.486 | 0.0294 | No | ||
| 13 | SLC25A12 | 1778 | 1.480 | 0.0370 | No | ||
| 14 | COL4A3 | 1836 | 1.460 | 0.0420 | No | ||
| 15 | IL19 | 2031 | 1.387 | 0.0392 | No | ||
| 16 | ASGR1 | 2116 | 1.360 | 0.0423 | No | ||
| 17 | ITPR3 | 2174 | 1.342 | 0.0467 | No | ||
| 18 | GOT1 | 2207 | 1.327 | 0.0524 | No | ||
| 19 | ANGPT1 | 2230 | 1.321 | 0.0585 | No | ||
| 20 | WNT5A | 2425 | 1.262 | 0.0551 | No | ||
| 21 | DBH | 2534 | 1.232 | 0.0561 | No | ||
| 22 | NDRG1 | 2571 | 1.221 | 0.0610 | No | ||
| 23 | CBX4 | 2630 | 1.208 | 0.0646 | No | ||
| 24 | CDKL5 | 2820 | 1.156 | 0.0608 | No | ||
| 25 | ARRDC3 | 3055 | 1.096 | 0.0542 | No | ||
| 26 | GLP2R | 3293 | 1.038 | 0.0471 | No | ||
| 27 | EDN2 | 3442 | 1.004 | 0.0447 | No | ||
| 28 | SMARCAD1 | 3708 | 0.942 | 0.0356 | No | ||
| 29 | NFE2L2 | 3720 | 0.939 | 0.0403 | No | ||
| 30 | MOSPD2 | 3770 | 0.930 | 0.0428 | No | ||
| 31 | SOX5 | 3879 | 0.906 | 0.0420 | No | ||
| 32 | FGF14 | 3897 | 0.900 | 0.0461 | No | ||
| 33 | NFIL3 | 4191 | 0.847 | 0.0349 | No | ||
| 34 | BCL11A | 4328 | 0.819 | 0.0321 | No | ||
| 35 | DLX1 | 4421 | 0.802 | 0.0316 | No | ||
| 36 | GYS1 | 4504 | 0.787 | 0.0316 | No | ||
| 37 | PDE4D | 4578 | 0.772 | 0.0319 | No | ||
| 38 | CUEDC1 | 4793 | 0.732 | 0.0244 | No | ||
| 39 | FGB | 4838 | 0.725 | 0.0261 | No | ||
| 40 | HOXC13 | 4844 | 0.724 | 0.0298 | No | ||
| 41 | PDE3B | 4858 | 0.721 | 0.0332 | No | ||
| 42 | CHRM1 | 5138 | 0.669 | 0.0218 | No | ||
| 43 | PRG4 | 5343 | 0.634 | 0.0142 | No | ||
| 44 | SULF1 | 5396 | 0.623 | 0.0149 | No | ||
| 45 | RIN3 | 5543 | 0.596 | 0.0103 | No | ||
| 46 | SMAD1 | 5622 | 0.584 | 0.0093 | No | ||
| 47 | PLA2G4A | 5724 | 0.569 | 0.0070 | No | ||
| 48 | SERPINA7 | 5831 | 0.552 | 0.0044 | No | ||
| 49 | CASK | 5835 | 0.552 | 0.0073 | No | ||
| 50 | TGFB3 | 6236 | 0.483 | -0.0117 | No | ||
| 51 | MYO1C | 6243 | 0.482 | -0.0093 | No | ||
| 52 | HOXA5 | 6345 | 0.463 | -0.0122 | No | ||
| 53 | ADRB2 | 6364 | 0.460 | -0.0106 | No | ||
| 54 | EIF4A2 | 6486 | 0.442 | -0.0147 | No | ||
| 55 | STAT3 | 6509 | 0.438 | -0.0135 | No | ||
| 56 | ALDH1A2 | 6583 | 0.426 | -0.0151 | No | ||
| 57 | PAFAH2 | 6696 | 0.408 | -0.0189 | No | ||
| 58 | ERF | 6871 | 0.378 | -0.0262 | No | ||
| 59 | MEOX2 | 6877 | 0.377 | -0.0243 | No | ||
| 60 | KCNJ2 | 7040 | 0.353 | -0.0312 | No | ||
| 61 | ZADH2 | 7041 | 0.353 | -0.0292 | No | ||
| 62 | EHF | 7176 | 0.330 | -0.0346 | No | ||
| 63 | RFX4 | 7378 | 0.296 | -0.0439 | No | ||
| 64 | SEPHS2 | 7500 | 0.278 | -0.0489 | No | ||
| 65 | C1QL1 | 7534 | 0.273 | -0.0492 | No | ||
| 66 | PHOX2B | 7550 | 0.271 | -0.0485 | No | ||
| 67 | RAD23B | 7702 | 0.247 | -0.0553 | No | ||
| 68 | RG9MTD2 | 7719 | 0.244 | -0.0548 | No | ||
| 69 | MRC2 | 7790 | 0.237 | -0.0572 | No | ||
| 70 | LEMD2 | 7829 | 0.232 | -0.0580 | No | ||
| 71 | PCF11 | 7863 | 0.227 | -0.0585 | No | ||
| 72 | SP8 | 7925 | 0.217 | -0.0606 | No | ||
| 73 | IL1RAPL2 | 7983 | 0.207 | -0.0626 | No | ||
| 74 | BMF | 8100 | 0.189 | -0.0678 | No | ||
| 75 | PDCL | 8102 | 0.189 | -0.0668 | No | ||
| 76 | NHLH1 | 8339 | 0.151 | -0.0788 | No | ||
| 77 | ELAVL2 | 8649 | 0.101 | -0.0949 | No | ||
| 78 | BNIP3L | 8965 | 0.053 | -0.1117 | No | ||
| 79 | SLC19A3 | 9183 | 0.021 | -0.1234 | No | ||
| 80 | BDKRB1 | 9468 | -0.022 | -0.1387 | No | ||
| 81 | CHD2 | 9588 | -0.042 | -0.1449 | No | ||
| 82 | RBPMS | 9866 | -0.092 | -0.1594 | No | ||
| 83 | RGR | 10221 | -0.145 | -0.1778 | No | ||
| 84 | MAP3K3 | 10229 | -0.147 | -0.1774 | No | ||
| 85 | PIP5K1A | 10312 | -0.158 | -0.1809 | No | ||
| 86 | SERPINF1 | 10452 | -0.179 | -0.1875 | No | ||
| 87 | KCNH7 | 10507 | -0.187 | -0.1894 | No | ||
| 88 | LMO4 | 10636 | -0.207 | -0.1951 | No | ||
| 89 | SDPR | 10659 | -0.210 | -0.1952 | No | ||
| 90 | FCER1G | 10964 | -0.263 | -0.2102 | No | ||
| 91 | PDGFRL | 11004 | -0.269 | -0.2108 | No | ||
| 92 | SPRY4 | 11338 | -0.318 | -0.2271 | No | ||
| 93 | CYP24A1 | 11372 | -0.322 | -0.2271 | No | ||
| 94 | IL27 | 11386 | -0.323 | -0.2260 | No | ||
| 95 | P2RY4 | 11633 | -0.355 | -0.2373 | No | ||
| 96 | PRDX3 | 11861 | -0.389 | -0.2475 | No | ||
| 97 | HIST1H1C | 11869 | -0.391 | -0.2457 | No | ||
| 98 | CP | 12001 | -0.409 | -0.2505 | No | ||
| 99 | ASAH2 | 12083 | -0.423 | -0.2525 | No | ||
| 100 | CSMD3 | 12138 | -0.432 | -0.2530 | No | ||
| 101 | PPP1CB | 12614 | -0.500 | -0.2760 | No | ||
| 102 | TMPRSS4 | 12624 | -0.502 | -0.2737 | No | ||
| 103 | ITGA11 | 12650 | -0.506 | -0.2722 | No | ||
| 104 | STX18 | 12775 | -0.526 | -0.2760 | No | ||
| 105 | ESR1 | 12798 | -0.528 | -0.2743 | No | ||
| 106 | PPL | 12906 | -0.550 | -0.2770 | No | ||
| 107 | PDGFB | 12928 | -0.553 | -0.2751 | No | ||
| 108 | H2AFZ | 13036 | -0.570 | -0.2777 | No | ||
| 109 | HAVCR1 | 13306 | -0.614 | -0.2888 | No | ||
| 110 | PPM1B | 13575 | -0.662 | -0.2997 | No | ||
| 111 | SMARCA1 | 13616 | -0.667 | -0.2981 | No | ||
| 112 | S100A9 | 13780 | -0.698 | -0.3031 | No | ||
| 113 | BCL6 | 13882 | -0.715 | -0.3046 | No | ||
| 114 | SMAD6 | 14046 | -0.743 | -0.3093 | No | ||
| 115 | TRIB1 | 14126 | -0.760 | -0.3093 | No | ||
| 116 | PCTP | 14402 | -0.810 | -0.3197 | No | ||
| 117 | CDKN1B | 14528 | -0.836 | -0.3218 | Yes | ||
| 118 | PTPN12 | 14599 | -0.850 | -0.3208 | Yes | ||
| 119 | DYRK3 | 14705 | -0.870 | -0.3217 | Yes | ||
| 120 | RPA3 | 14709 | -0.872 | -0.3170 | Yes | ||
| 121 | RPS21 | 14782 | -0.887 | -0.3159 | Yes | ||
| 122 | AQP9 | 14915 | -0.915 | -0.3180 | Yes | ||
| 123 | MBNL1 | 14932 | -0.919 | -0.3137 | Yes | ||
| 124 | TOB1 | 14949 | -0.923 | -0.3094 | Yes | ||
| 125 | KCNE3 | 14954 | -0.924 | -0.3045 | Yes | ||
| 126 | LEP | 14973 | -0.929 | -0.3003 | Yes | ||
| 127 | SPRED1 | 15111 | -0.955 | -0.3024 | Yes | ||
| 128 | UGP2 | 15190 | -0.969 | -0.3012 | Yes | ||
| 129 | MAP4K4 | 15211 | -0.972 | -0.2968 | Yes | ||
| 130 | FST | 15277 | -0.988 | -0.2949 | Yes | ||
| 131 | FMO2 | 15430 | -1.020 | -0.2974 | Yes | ||
| 132 | ASCL2 | 15534 | -1.041 | -0.2972 | Yes | ||
| 133 | PDAP1 | 15535 | -1.041 | -0.2914 | Yes | ||
| 134 | MAP2K6 | 15559 | -1.047 | -0.2868 | Yes | ||
| 135 | PHF16 | 15603 | -1.057 | -0.2832 | Yes | ||
| 136 | TCF12 | 15625 | -1.060 | -0.2784 | Yes | ||
| 137 | SERTAD4 | 15665 | -1.067 | -0.2746 | Yes | ||
| 138 | PFN2 | 15681 | -1.071 | -0.2694 | Yes | ||
| 139 | MPP6 | 15703 | -1.075 | -0.2645 | Yes | ||
| 140 | SLC7A11 | 15826 | -1.105 | -0.2650 | Yes | ||
| 141 | CCL3 | 15932 | -1.138 | -0.2643 | Yes | ||
| 142 | UBE2E2 | 16086 | -1.174 | -0.2660 | Yes | ||
| 143 | ARNT | 16099 | -1.176 | -0.2601 | Yes | ||
| 144 | CITED1 | 16214 | -1.206 | -0.2596 | Yes | ||
| 145 | TBR1 | 16226 | -1.210 | -0.2534 | Yes | ||
| 146 | RUVBL2 | 16269 | -1.224 | -0.2488 | Yes | ||
| 147 | THRA | 16282 | -1.228 | -0.2426 | Yes | ||
| 148 | RHOB | 16287 | -1.229 | -0.2360 | Yes | ||
| 149 | ATAD2 | 16327 | -1.238 | -0.2312 | Yes | ||
| 150 | ARF6 | 16458 | -1.269 | -0.2312 | Yes | ||
| 151 | G0S2 | 16482 | -1.275 | -0.2253 | Yes | ||
| 152 | BNC2 | 16494 | -1.280 | -0.2187 | Yes | ||
| 153 | RRBP1 | 16522 | -1.288 | -0.2130 | Yes | ||
| 154 | ACLY | 16634 | -1.320 | -0.2116 | Yes | ||
| 155 | EIF4A1 | 16719 | -1.349 | -0.2087 | Yes | ||
| 156 | EFNA5 | 16760 | -1.361 | -0.2032 | Yes | ||
| 157 | PDGFRA | 16894 | -1.401 | -0.2026 | Yes | ||
| 158 | NFKBIA | 16992 | -1.433 | -0.1999 | Yes | ||
| 159 | SLC12A1 | 17006 | -1.438 | -0.1926 | Yes | ||
| 160 | KPNA3 | 17033 | -1.448 | -0.1859 | Yes | ||
| 161 | SLIT3 | 17104 | -1.478 | -0.1814 | Yes | ||
| 162 | HNRPR | 17112 | -1.479 | -0.1735 | Yes | ||
| 163 | CACNB2 | 17265 | -1.546 | -0.1731 | Yes | ||
| 164 | MTF1 | 17273 | -1.548 | -0.1649 | Yes | ||
| 165 | PPARG | 17277 | -1.551 | -0.1564 | Yes | ||
| 166 | CSNK1E | 17374 | -1.594 | -0.1527 | Yes | ||
| 167 | ONECUT2 | 17405 | -1.608 | -0.1453 | Yes | ||
| 168 | DMD | 17599 | -1.693 | -0.1463 | Yes | ||
| 169 | LUZP1 | 17667 | -1.726 | -0.1403 | Yes | ||
| 170 | SOCS2 | 17815 | -1.816 | -0.1382 | Yes | ||
| 171 | SYNCRIP | 17816 | -1.816 | -0.1280 | Yes | ||
| 172 | FIGN | 17891 | -1.869 | -0.1216 | Yes | ||
| 173 | H3F3B | 17921 | -1.889 | -0.1126 | Yes | ||
| 174 | TOP1 | 18142 | -2.093 | -0.1129 | Yes | ||
| 175 | CCND2 | 18144 | -2.098 | -0.1012 | Yes | ||
| 176 | FGA | 18237 | -2.213 | -0.0938 | Yes | ||
| 177 | NEK6 | 18260 | -2.236 | -0.0825 | Yes | ||
| 178 | SLC6A4 | 18273 | -2.268 | -0.0705 | Yes | ||
| 179 | FOXP1 | 18307 | -2.323 | -0.0593 | Yes | ||
| 180 | VNN3 | 18371 | -2.413 | -0.0493 | Yes | ||
| 181 | PER1 | 18385 | -2.454 | -0.0363 | Yes | ||
| 182 | DUSP1 | 18504 | -2.834 | -0.0268 | Yes | ||
| 183 | PCDH7 | 18515 | -2.871 | -0.0113 | Yes | ||
| 184 | UBQLN1 | 18541 | -3.000 | 0.0041 | Yes |