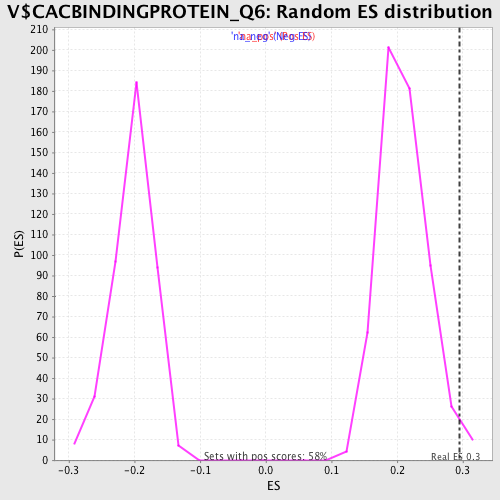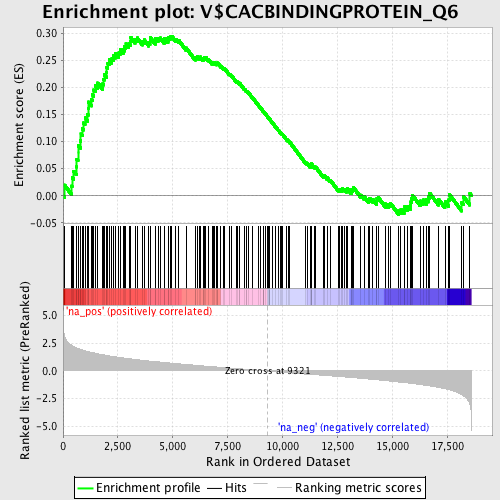
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$CACBINDINGPROTEIN_Q6 |
| Enrichment Score (ES) | 0.2944888 |
| Normalized Enrichment Score (NES) | 1.4030975 |
| Nominal p-value | 0.017271157 |
| FDR q-value | 0.49920934 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AAMP | 58 | 3.218 | 0.0198 | Yes | ||
| 2 | FBXO36 | 397 | 2.311 | 0.0180 | Yes | ||
| 3 | CACNA1G | 411 | 2.289 | 0.0336 | Yes | ||
| 4 | GRK5 | 483 | 2.207 | 0.0455 | Yes | ||
| 5 | HOXC6 | 614 | 2.076 | 0.0533 | Yes | ||
| 6 | PKIA | 623 | 2.070 | 0.0677 | Yes | ||
| 7 | HOXB6 | 681 | 2.018 | 0.0790 | Yes | ||
| 8 | ACR | 682 | 2.017 | 0.0934 | Yes | ||
| 9 | KCNJ14 | 787 | 1.948 | 0.1016 | Yes | ||
| 10 | OTX2 | 814 | 1.935 | 0.1140 | Yes | ||
| 11 | GNGT2 | 881 | 1.889 | 0.1239 | Yes | ||
| 12 | ADRA1D | 934 | 1.855 | 0.1344 | Yes | ||
| 13 | FOXA2 | 1000 | 1.817 | 0.1438 | Yes | ||
| 14 | FBS1 | 1120 | 1.753 | 0.1499 | Yes | ||
| 15 | TP53 | 1142 | 1.741 | 0.1612 | Yes | ||
| 16 | MKL1 | 1153 | 1.737 | 0.1730 | Yes | ||
| 17 | MTX1 | 1292 | 1.676 | 0.1775 | Yes | ||
| 18 | ROCK1 | 1338 | 1.656 | 0.1869 | Yes | ||
| 19 | SALL1 | 1401 | 1.632 | 0.1952 | Yes | ||
| 20 | ARF3 | 1467 | 1.608 | 0.2031 | Yes | ||
| 21 | AP1G1 | 1582 | 1.558 | 0.2081 | Yes | ||
| 22 | GNB4 | 1808 | 1.470 | 0.2064 | Yes | ||
| 23 | ARHGEF17 | 1849 | 1.456 | 0.2146 | Yes | ||
| 24 | ZSWIM3 | 1872 | 1.446 | 0.2237 | Yes | ||
| 25 | EFNB3 | 1980 | 1.404 | 0.2279 | Yes | ||
| 26 | DTNA | 1991 | 1.401 | 0.2374 | Yes | ||
| 27 | PRRX1 | 2038 | 1.384 | 0.2448 | Yes | ||
| 28 | SIX4 | 2101 | 1.366 | 0.2512 | Yes | ||
| 29 | HR | 2225 | 1.322 | 0.2539 | Yes | ||
| 30 | TRIM46 | 2305 | 1.298 | 0.2589 | Yes | ||
| 31 | DCTN2 | 2397 | 1.270 | 0.2630 | Yes | ||
| 32 | ATP6V0C | 2546 | 1.228 | 0.2638 | Yes | ||
| 33 | EEF1A1 | 2598 | 1.216 | 0.2697 | Yes | ||
| 34 | ACIN1 | 2737 | 1.180 | 0.2706 | Yes | ||
| 35 | NOTCH2 | 2808 | 1.158 | 0.2751 | Yes | ||
| 36 | KLK12 | 2865 | 1.146 | 0.2803 | Yes | ||
| 37 | ZFYVE1 | 3003 | 1.109 | 0.2807 | Yes | ||
| 38 | FGF11 | 3065 | 1.094 | 0.2852 | Yes | ||
| 39 | NEGR1 | 3086 | 1.088 | 0.2919 | Yes | ||
| 40 | ACVR1B | 3278 | 1.042 | 0.2890 | Yes | ||
| 41 | STRN3 | 3372 | 1.020 | 0.2913 | Yes | ||
| 42 | RFX1 | 3617 | 0.963 | 0.2849 | Yes | ||
| 43 | AP4S1 | 3691 | 0.948 | 0.2877 | Yes | ||
| 44 | KCNN3 | 3907 | 0.899 | 0.2825 | Yes | ||
| 45 | HIF1A | 3962 | 0.887 | 0.2859 | Yes | ||
| 46 | EEF1G | 3969 | 0.885 | 0.2919 | Yes | ||
| 47 | NCDN | 4196 | 0.846 | 0.2857 | Yes | ||
| 48 | RNF12 | 4211 | 0.842 | 0.2909 | Yes | ||
| 49 | BCL11A | 4328 | 0.819 | 0.2905 | Yes | ||
| 50 | DLX1 | 4421 | 0.802 | 0.2912 | Yes | ||
| 51 | PHF12 | 4604 | 0.767 | 0.2868 | Yes | ||
| 52 | DCN | 4631 | 0.760 | 0.2909 | Yes | ||
| 53 | PCTK2 | 4782 | 0.734 | 0.2880 | Yes | ||
| 54 | CACNA1D | 4810 | 0.730 | 0.2917 | Yes | ||
| 55 | TNNC1 | 4877 | 0.718 | 0.2933 | Yes | ||
| 56 | KRTCAP2 | 4948 | 0.705 | 0.2945 | Yes | ||
| 57 | CHRM1 | 5138 | 0.669 | 0.2890 | No | ||
| 58 | FKHL18 | 5267 | 0.645 | 0.2867 | No | ||
| 59 | BCL7A | 5604 | 0.587 | 0.2727 | No | ||
| 60 | ADAMTS4 | 6019 | 0.520 | 0.2539 | No | ||
| 61 | HEBP2 | 6053 | 0.513 | 0.2558 | No | ||
| 62 | DPF1 | 6103 | 0.504 | 0.2568 | No | ||
| 63 | NR4A3 | 6198 | 0.489 | 0.2552 | No | ||
| 64 | MARK2 | 6244 | 0.482 | 0.2562 | No | ||
| 65 | INPP4A | 6384 | 0.457 | 0.2519 | No | ||
| 66 | NTF5 | 6410 | 0.453 | 0.2538 | No | ||
| 67 | APOA2 | 6433 | 0.448 | 0.2558 | No | ||
| 68 | ARHGEF12 | 6504 | 0.439 | 0.2551 | No | ||
| 69 | ARPC2 | 6629 | 0.418 | 0.2514 | No | ||
| 70 | HOXC4 | 6793 | 0.391 | 0.2453 | No | ||
| 71 | RALB | 6859 | 0.380 | 0.2445 | No | ||
| 72 | MAGED2 | 6883 | 0.376 | 0.2460 | No | ||
| 73 | RAB6B | 6972 | 0.362 | 0.2438 | No | ||
| 74 | HPCAL4 | 7016 | 0.356 | 0.2440 | No | ||
| 75 | ZADH2 | 7041 | 0.353 | 0.2452 | No | ||
| 76 | TITF1 | 7189 | 0.328 | 0.2396 | No | ||
| 77 | GDPD2 | 7311 | 0.307 | 0.2352 | No | ||
| 78 | UBE2B | 7349 | 0.300 | 0.2353 | No | ||
| 79 | SUPT16H | 7584 | 0.265 | 0.2245 | No | ||
| 80 | IVD | 7681 | 0.250 | 0.2211 | No | ||
| 81 | HOXB8 | 7908 | 0.220 | 0.2104 | No | ||
| 82 | PHACTR3 | 7938 | 0.215 | 0.2104 | No | ||
| 83 | ABI3 | 8033 | 0.198 | 0.2067 | No | ||
| 84 | ADAMTS9 | 8036 | 0.197 | 0.2080 | No | ||
| 85 | NEDD4L | 8266 | 0.165 | 0.1968 | No | ||
| 86 | EMX2 | 8354 | 0.150 | 0.1931 | No | ||
| 87 | VPS35 | 8376 | 0.147 | 0.1931 | No | ||
| 88 | PRKAG1 | 8468 | 0.132 | 0.1891 | No | ||
| 89 | THBS3 | 8625 | 0.105 | 0.1814 | No | ||
| 90 | NRGN | 8888 | 0.063 | 0.1676 | No | ||
| 91 | CASKIN2 | 8988 | 0.050 | 0.1626 | No | ||
| 92 | NOL4 | 9136 | 0.029 | 0.1548 | No | ||
| 93 | KNTC1 | 9137 | 0.029 | 0.1551 | No | ||
| 94 | EIF4G1 | 9140 | 0.028 | 0.1551 | No | ||
| 95 | FZD4 | 9209 | 0.018 | 0.1516 | No | ||
| 96 | PROCR | 9312 | 0.002 | 0.1461 | No | ||
| 97 | MTHFR | 9348 | -0.005 | 0.1442 | No | ||
| 98 | NXPH3 | 9396 | -0.012 | 0.1417 | No | ||
| 99 | NMT1 | 9405 | -0.014 | 0.1414 | No | ||
| 100 | IGFBP5 | 9408 | -0.014 | 0.1414 | No | ||
| 101 | LIN28 | 9535 | -0.032 | 0.1348 | No | ||
| 102 | MGAT3 | 9689 | -0.060 | 0.1269 | No | ||
| 103 | KIF1C | 9824 | -0.085 | 0.1203 | No | ||
| 104 | MYH10 | 9894 | -0.098 | 0.1173 | No | ||
| 105 | RTN4RL2 | 9958 | -0.109 | 0.1146 | No | ||
| 106 | PABPN1 | 9992 | -0.113 | 0.1136 | No | ||
| 107 | S100A14 | 10198 | -0.143 | 0.1035 | No | ||
| 108 | CLCN6 | 10260 | -0.151 | 0.1013 | No | ||
| 109 | PICALM | 10336 | -0.162 | 0.0984 | No | ||
| 110 | KCNS3 | 11056 | -0.277 | 0.0614 | No | ||
| 111 | DCTN1 | 11123 | -0.287 | 0.0599 | No | ||
| 112 | HTATIP | 11272 | -0.308 | 0.0541 | No | ||
| 113 | KLF13 | 11302 | -0.313 | 0.0547 | No | ||
| 114 | PYGO1 | 11303 | -0.313 | 0.0570 | No | ||
| 115 | MYBPH | 11304 | -0.313 | 0.0592 | No | ||
| 116 | MEF2D | 11458 | -0.331 | 0.0533 | No | ||
| 117 | ACVRL1 | 11506 | -0.338 | 0.0531 | No | ||
| 118 | RALBP1 | 11881 | -0.393 | 0.0357 | No | ||
| 119 | SLC2A4 | 11897 | -0.395 | 0.0377 | No | ||
| 120 | USF2 | 12031 | -0.414 | 0.0334 | No | ||
| 121 | ITGA6 | 12181 | -0.438 | 0.0285 | No | ||
| 122 | KIF3C | 12552 | -0.491 | 0.0119 | No | ||
| 123 | PPP1CB | 12614 | -0.500 | 0.0122 | No | ||
| 124 | AKT2 | 12698 | -0.515 | 0.0114 | No | ||
| 125 | SDC1 | 12730 | -0.519 | 0.0134 | No | ||
| 126 | MNT | 12820 | -0.534 | 0.0124 | No | ||
| 127 | PDGFB | 12928 | -0.553 | 0.0105 | No | ||
| 128 | HOXB9 | 12954 | -0.557 | 0.0131 | No | ||
| 129 | KPNB1 | 13132 | -0.583 | 0.0077 | No | ||
| 130 | CSDA | 13145 | -0.585 | 0.0112 | No | ||
| 131 | ARHGAP5 | 13207 | -0.595 | 0.0122 | No | ||
| 132 | AHR | 13223 | -0.597 | 0.0156 | No | ||
| 133 | JPH4 | 13574 | -0.661 | 0.0014 | No | ||
| 134 | RAB11B | 13719 | -0.687 | -0.0015 | No | ||
| 135 | LRP1 | 13928 | -0.723 | -0.0076 | No | ||
| 136 | NR1H3 | 13970 | -0.730 | -0.0046 | No | ||
| 137 | YWHAQ | 14103 | -0.753 | -0.0064 | No | ||
| 138 | LRFN5 | 14289 | -0.791 | -0.0108 | No | ||
| 139 | HOXA10 | 14301 | -0.794 | -0.0057 | No | ||
| 140 | RAB6A | 14352 | -0.803 | -0.0027 | No | ||
| 141 | ZNF385 | 14693 | -0.868 | -0.0149 | No | ||
| 142 | MAP1A | 14824 | -0.896 | -0.0156 | No | ||
| 143 | SYT6 | 14907 | -0.914 | -0.0135 | No | ||
| 144 | PIP5K2B | 15297 | -0.992 | -0.0275 | No | ||
| 145 | RTN2 | 15397 | -1.013 | -0.0257 | No | ||
| 146 | PBX1 | 15541 | -1.043 | -0.0260 | No | ||
| 147 | LASP1 | 15574 | -1.050 | -0.0202 | No | ||
| 148 | KIRREL2 | 15713 | -1.079 | -0.0200 | No | ||
| 149 | GNAO1 | 15824 | -1.105 | -0.0180 | No | ||
| 150 | PIM1 | 15850 | -1.112 | -0.0115 | No | ||
| 151 | NEUROD2 | 15880 | -1.124 | -0.0050 | No | ||
| 152 | POU2F1 | 15913 | -1.133 | 0.0013 | No | ||
| 153 | NDUFS2 | 16277 | -1.227 | -0.0096 | No | ||
| 154 | ATF3 | 16408 | -1.257 | -0.0077 | No | ||
| 155 | ORC6L | 16565 | -1.299 | -0.0068 | No | ||
| 156 | EMID1 | 16654 | -1.327 | -0.0021 | No | ||
| 157 | LRP2 | 16702 | -1.343 | 0.0049 | No | ||
| 158 | HNRPR | 17112 | -1.479 | -0.0067 | No | ||
| 159 | CHL1 | 17406 | -1.609 | -0.0111 | No | ||
| 160 | NFIX | 17566 | -1.676 | -0.0078 | No | ||
| 161 | DMD | 17599 | -1.693 | 0.0026 | No | ||
| 162 | ITPR1 | 18169 | -2.124 | -0.0131 | No | ||
| 163 | NCAM1 | 18242 | -2.217 | -0.0012 | No | ||
| 164 | EPC1 | 18540 | -2.994 | 0.0041 | No |