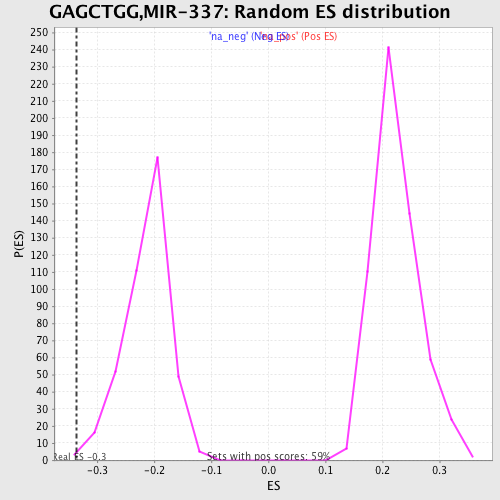
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GAGCTGG,MIR-337 |
| Enrichment Score (ES) | -0.33695924 |
| Normalized Enrichment Score (NES) | -1.5858719 |
| Nominal p-value | 0.0072639226 |
| FDR q-value | 0.29782054 |
| FWER p-Value | 0.817 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | NFASC | 347 | 2.376 | 0.0049 | No | ||
| 2 | VAMP1 | 521 | 2.168 | 0.0171 | No | ||
| 3 | EI24 | 935 | 1.855 | 0.0133 | No | ||
| 4 | DGKZ | 1278 | 1.680 | 0.0115 | No | ||
| 5 | SEMA6A | 1284 | 1.679 | 0.0279 | No | ||
| 6 | PPP1R9B | 1543 | 1.577 | 0.0297 | No | ||
| 7 | TOR2A | 1865 | 1.449 | 0.0268 | No | ||
| 8 | PDGFRB | 2074 | 1.375 | 0.0292 | No | ||
| 9 | TRIM35 | 2359 | 1.281 | 0.0266 | No | ||
| 10 | KCNH8 | 2584 | 1.218 | 0.0266 | No | ||
| 11 | JUNB | 2898 | 1.135 | 0.0210 | No | ||
| 12 | FRAG1 | 2912 | 1.131 | 0.0316 | No | ||
| 13 | XBP1 | 2913 | 1.131 | 0.0428 | No | ||
| 14 | SLC25A22 | 3246 | 1.049 | 0.0353 | No | ||
| 15 | LBX1 | 3260 | 1.045 | 0.0450 | No | ||
| 16 | LRRC4 | 3501 | 0.991 | 0.0419 | No | ||
| 17 | FBXL17 | 4235 | 0.837 | 0.0107 | No | ||
| 18 | MAMDC1 | 4445 | 0.798 | 0.0073 | No | ||
| 19 | ROM1 | 4450 | 0.798 | 0.0150 | No | ||
| 20 | PHF13 | 5165 | 0.664 | -0.0169 | No | ||
| 21 | EHMT2 | 5195 | 0.659 | -0.0119 | No | ||
| 22 | LIMK2 | 5241 | 0.650 | -0.0079 | No | ||
| 23 | PPP1R3F | 5632 | 0.583 | -0.0232 | No | ||
| 24 | RNF121 | 6090 | 0.505 | -0.0428 | No | ||
| 25 | B3GAT3 | 6253 | 0.481 | -0.0468 | No | ||
| 26 | BCAP29 | 6325 | 0.467 | -0.0460 | No | ||
| 27 | PLAGL2 | 6326 | 0.467 | -0.0413 | No | ||
| 28 | TEAD3 | 6563 | 0.429 | -0.0498 | No | ||
| 29 | ALDH1A2 | 6583 | 0.426 | -0.0466 | No | ||
| 30 | MCRS1 | 6622 | 0.420 | -0.0445 | No | ||
| 31 | POGZ | 7141 | 0.336 | -0.0691 | No | ||
| 32 | KLF11 | 7591 | 0.264 | -0.0908 | No | ||
| 33 | TXNL4B | 7808 | 0.235 | -0.1001 | No | ||
| 34 | SEC24C | 7834 | 0.231 | -0.0991 | No | ||
| 35 | SP7 | 8039 | 0.197 | -0.1082 | No | ||
| 36 | RANBP10 | 8125 | 0.187 | -0.1109 | No | ||
| 37 | FOSL1 | 8533 | 0.121 | -0.1317 | No | ||
| 38 | IER5L | 8687 | 0.095 | -0.1390 | No | ||
| 39 | VCP | 9030 | 0.044 | -0.1571 | No | ||
| 40 | CAMK2G | 9064 | 0.040 | -0.1585 | No | ||
| 41 | HOXB5 | 9351 | -0.006 | -0.1739 | No | ||
| 42 | RARA | 9352 | -0.006 | -0.1738 | No | ||
| 43 | RGS16 | 9371 | -0.009 | -0.1747 | No | ||
| 44 | TWIST1 | 9443 | -0.019 | -0.1783 | No | ||
| 45 | PHF2 | 9678 | -0.057 | -0.1904 | No | ||
| 46 | MAP3K9 | 9734 | -0.068 | -0.1927 | No | ||
| 47 | PARVA | 9931 | -0.103 | -0.2023 | No | ||
| 48 | MAP3K3 | 10229 | -0.147 | -0.2168 | No | ||
| 49 | DNAJB5 | 10626 | -0.206 | -0.2362 | No | ||
| 50 | FOS | 10648 | -0.208 | -0.2353 | No | ||
| 51 | BCL2L1 | 10664 | -0.211 | -0.2340 | No | ||
| 52 | STIP1 | 11074 | -0.279 | -0.2533 | No | ||
| 53 | KLF13 | 11302 | -0.313 | -0.2624 | No | ||
| 54 | SSBP3 | 11454 | -0.330 | -0.2673 | No | ||
| 55 | RNF19 | 11649 | -0.357 | -0.2742 | No | ||
| 56 | DPH2 | 11916 | -0.398 | -0.2846 | No | ||
| 57 | SLC12A5 | 11928 | -0.399 | -0.2813 | No | ||
| 58 | SMARCD2 | 12398 | -0.467 | -0.3020 | No | ||
| 59 | CHRD | 12458 | -0.476 | -0.3004 | No | ||
| 60 | TYSND1 | 12579 | -0.495 | -0.3020 | No | ||
| 61 | ITGA11 | 12650 | -0.506 | -0.3007 | No | ||
| 62 | GPD1 | 12982 | -0.561 | -0.3130 | No | ||
| 63 | BRD2 | 13136 | -0.583 | -0.3155 | No | ||
| 64 | B4GALT3 | 13305 | -0.613 | -0.3185 | No | ||
| 65 | RASGRP4 | 13338 | -0.619 | -0.3140 | No | ||
| 66 | SLC1A2 | 13367 | -0.625 | -0.3093 | No | ||
| 67 | MRPS16 | 13424 | -0.635 | -0.3060 | No | ||
| 68 | UBAP1 | 13454 | -0.640 | -0.3012 | No | ||
| 69 | SFRS1 | 13638 | -0.672 | -0.3044 | No | ||
| 70 | LPHN1 | 13834 | -0.709 | -0.3079 | No | ||
| 71 | KCND1 | 13947 | -0.727 | -0.3067 | No | ||
| 72 | PARP6 | 13993 | -0.733 | -0.3019 | No | ||
| 73 | AP1S3 | 14174 | -0.769 | -0.3039 | No | ||
| 74 | BNIPL | 14217 | -0.775 | -0.2985 | No | ||
| 75 | SBK1 | 14400 | -0.809 | -0.3003 | No | ||
| 76 | SEPT6 | 14923 | -0.917 | -0.3193 | No | ||
| 77 | GRK6 | 15250 | -0.978 | -0.3272 | Yes | ||
| 78 | GPRASP2 | 15299 | -0.992 | -0.3199 | Yes | ||
| 79 | ZMYND11 | 15466 | -1.028 | -0.3187 | Yes | ||
| 80 | ABCA9 | 15543 | -1.043 | -0.3124 | Yes | ||
| 81 | DSCAM | 15629 | -1.061 | -0.3064 | Yes | ||
| 82 | ACOT7 | 15688 | -1.072 | -0.2989 | Yes | ||
| 83 | MAT2A | 16104 | -1.177 | -0.3096 | Yes | ||
| 84 | ZFAND3 | 16175 | -1.196 | -0.3015 | Yes | ||
| 85 | THRA | 16282 | -1.228 | -0.2950 | Yes | ||
| 86 | MFN1 | 16319 | -1.237 | -0.2847 | Yes | ||
| 87 | RNF44 | 16410 | -1.257 | -0.2770 | Yes | ||
| 88 | BTG2 | 16426 | -1.261 | -0.2653 | Yes | ||
| 89 | BNC2 | 16494 | -1.280 | -0.2562 | Yes | ||
| 90 | RAB15 | 16549 | -1.295 | -0.2462 | Yes | ||
| 91 | ABCF1 | 16571 | -1.302 | -0.2344 | Yes | ||
| 92 | COMMD3 | 16815 | -1.377 | -0.2338 | Yes | ||
| 93 | CHD4 | 16895 | -1.401 | -0.2241 | Yes | ||
| 94 | JPH1 | 16961 | -1.422 | -0.2135 | Yes | ||
| 95 | UHRF2 | 17281 | -1.552 | -0.2153 | Yes | ||
| 96 | EFNB1 | 17927 | -1.892 | -0.2313 | Yes | ||
| 97 | GLCCI1 | 17966 | -1.918 | -0.2143 | Yes | ||
| 98 | H1F0 | 18207 | -2.177 | -0.2056 | Yes | ||
| 99 | KLF12 | 18302 | -2.309 | -0.1877 | Yes | ||
| 100 | YPEL1 | 18326 | -2.346 | -0.1656 | Yes | ||
| 101 | NAV1 | 18355 | -2.387 | -0.1433 | Yes | ||
| 102 | PACSIN2 | 18380 | -2.427 | -0.1205 | Yes | ||
| 103 | TOMM34 | 18398 | -2.477 | -0.0967 | Yes | ||
| 104 | OVOL1 | 18427 | -2.541 | -0.0730 | Yes | ||
| 105 | NR1D1 | 18454 | -2.653 | -0.0480 | Yes | ||
| 106 | CD44 | 18469 | -2.701 | -0.0219 | Yes | ||
| 107 | SORT1 | 18539 | -2.990 | 0.0042 | Yes |