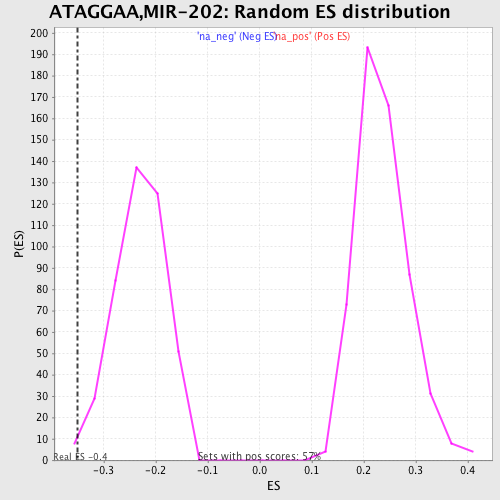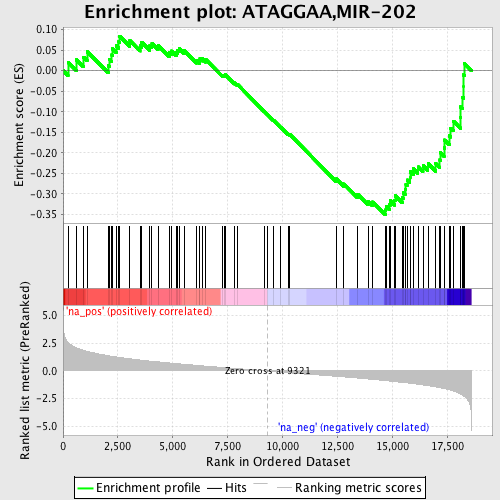
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATAGGAA,MIR-202 |
| Enrichment Score (ES) | -0.35008058 |
| Normalized Enrichment Score (NES) | -1.5127506 |
| Nominal p-value | 0.0069124424 |
| FDR q-value | 0.20794812 |
| FWER p-Value | 0.966 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ERBB2IP | 226 | 2.574 | 0.0208 | No | ||
| 2 | BCL2 | 612 | 2.077 | 0.0266 | No | ||
| 3 | EI24 | 935 | 1.855 | 0.0330 | No | ||
| 4 | SORCS1 | 1095 | 1.762 | 0.0470 | No | ||
| 5 | EAF1 | 2072 | 1.375 | 0.0120 | No | ||
| 6 | HIP2 | 2113 | 1.360 | 0.0272 | No | ||
| 7 | NARG1 | 2224 | 1.322 | 0.0382 | No | ||
| 8 | PCGF2 | 2257 | 1.312 | 0.0533 | No | ||
| 9 | BTG1 | 2420 | 1.264 | 0.0607 | No | ||
| 10 | ELF1 | 2540 | 1.229 | 0.0701 | No | ||
| 11 | NDRG1 | 2571 | 1.221 | 0.0841 | No | ||
| 12 | RASA1 | 3042 | 1.098 | 0.0728 | No | ||
| 13 | USP15 | 3506 | 0.989 | 0.0605 | No | ||
| 14 | KHDRBS2 | 3574 | 0.972 | 0.0693 | No | ||
| 15 | RKHD3 | 3954 | 0.890 | 0.0603 | No | ||
| 16 | GRIA3 | 4049 | 0.871 | 0.0664 | No | ||
| 17 | BCL11A | 4328 | 0.819 | 0.0619 | No | ||
| 18 | NR2F2 | 4837 | 0.726 | 0.0438 | No | ||
| 19 | KIAA1715 | 4929 | 0.709 | 0.0479 | No | ||
| 20 | TGFBR2 | 5156 | 0.666 | 0.0443 | No | ||
| 21 | SNX16 | 5217 | 0.655 | 0.0494 | No | ||
| 22 | VANGL2 | 5291 | 0.642 | 0.0537 | No | ||
| 23 | RAB22A | 5517 | 0.601 | 0.0493 | No | ||
| 24 | RNF121 | 6090 | 0.505 | 0.0249 | No | ||
| 25 | FAM60A | 6226 | 0.485 | 0.0238 | No | ||
| 26 | MARCKS | 6237 | 0.483 | 0.0295 | No | ||
| 27 | TRIM33 | 6350 | 0.463 | 0.0293 | No | ||
| 28 | STAT3 | 6509 | 0.438 | 0.0264 | No | ||
| 29 | PTEN | 7279 | 0.311 | -0.0110 | No | ||
| 30 | RPS6KA3 | 7352 | 0.299 | -0.0111 | No | ||
| 31 | BAG4 | 7386 | 0.295 | -0.0091 | No | ||
| 32 | HOXB2 | 7822 | 0.233 | -0.0296 | No | ||
| 33 | PPP5C | 7952 | 0.212 | -0.0338 | No | ||
| 34 | BICD2 | 9160 | 0.025 | -0.0986 | No | ||
| 35 | CPEB3 | 9327 | -0.002 | -0.1075 | No | ||
| 36 | IQGAP1 | 9599 | -0.043 | -0.1216 | No | ||
| 37 | SFPQ | 9917 | -0.101 | -0.1374 | No | ||
| 38 | TARDBP | 10273 | -0.153 | -0.1546 | No | ||
| 39 | MIB1 | 10321 | -0.159 | -0.1551 | No | ||
| 40 | HLF | 12442 | -0.474 | -0.2633 | No | ||
| 41 | ESR1 | 12798 | -0.528 | -0.2757 | No | ||
| 42 | ROBO2 | 13427 | -0.635 | -0.3014 | No | ||
| 43 | CNN3 | 13899 | -0.717 | -0.3177 | No | ||
| 44 | KRAS | 14116 | -0.757 | -0.3196 | No | ||
| 45 | TTC13 | 14682 | -0.865 | -0.3390 | Yes | ||
| 46 | USP8 | 14716 | -0.874 | -0.3296 | Yes | ||
| 47 | ACVR1 | 14876 | -0.909 | -0.3265 | Yes | ||
| 48 | DNAJC10 | 14904 | -0.913 | -0.3163 | Yes | ||
| 49 | SPRED1 | 15111 | -0.955 | -0.3152 | Yes | ||
| 50 | CREBBP | 15138 | -0.959 | -0.3043 | Yes | ||
| 51 | ANP32E | 15456 | -1.026 | -0.3083 | Yes | ||
| 52 | ARSB | 15500 | -1.034 | -0.2973 | Yes | ||
| 53 | ATXN1 | 15589 | -1.054 | -0.2886 | Yes | ||
| 54 | TCF12 | 15625 | -1.060 | -0.2769 | Yes | ||
| 55 | DDX3X | 15680 | -1.071 | -0.2661 | Yes | ||
| 56 | THRAP2 | 15810 | -1.102 | -0.2589 | Yes | ||
| 57 | SENP1 | 15818 | -1.103 | -0.2452 | Yes | ||
| 58 | SNAP91 | 15970 | -1.148 | -0.2386 | Yes | ||
| 59 | ZFAND3 | 16175 | -1.196 | -0.2343 | Yes | ||
| 60 | CD28 | 16403 | -1.256 | -0.2305 | Yes | ||
| 61 | ACSL3 | 16630 | -1.319 | -0.2258 | Yes | ||
| 62 | APPBP2 | 16985 | -1.431 | -0.2265 | Yes | ||
| 63 | YAF2 | 17173 | -1.504 | -0.2174 | Yes | ||
| 64 | MAPK6 | 17202 | -1.518 | -0.1994 | Yes | ||
| 65 | CTBP2 | 17366 | -1.590 | -0.1878 | Yes | ||
| 66 | PPARBP | 17392 | -1.601 | -0.1687 | Yes | ||
| 67 | LBR | 17604 | -1.696 | -0.1583 | Yes | ||
| 68 | LUZP1 | 17667 | -1.726 | -0.1396 | Yes | ||
| 69 | CBL | 17780 | -1.792 | -0.1227 | Yes | ||
| 70 | RNF11 | 18108 | -2.058 | -0.1140 | Yes | ||
| 71 | RAD52 | 18120 | -2.068 | -0.0881 | Yes | ||
| 72 | SSBP2 | 18206 | -2.176 | -0.0648 | Yes | ||
| 73 | ENAH | 18250 | -2.231 | -0.0385 | Yes | ||
| 74 | DDEF1 | 18254 | -2.232 | -0.0101 | Yes | ||
| 75 | KLF12 | 18302 | -2.309 | 0.0169 | Yes |