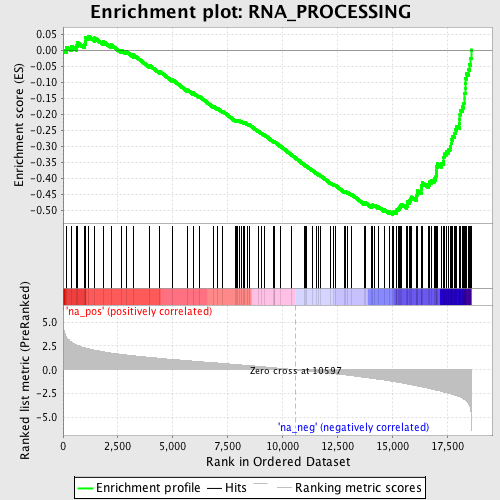
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | -0.513207 |
| Normalized Enrichment Score (NES) | -2.4450765 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.6297453E-4 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | METTL1 | 149 | 3.389 | 0.0093 | No | ||
| 2 | GEMIN7 | 367 | 2.923 | 0.0125 | No | ||
| 3 | SFRS8 | 597 | 2.619 | 0.0136 | No | ||
| 4 | SF3A2 | 668 | 2.545 | 0.0228 | No | ||
| 5 | IGF2BP3 | 965 | 2.304 | 0.0186 | No | ||
| 6 | SNRPD2 | 1010 | 2.276 | 0.0279 | No | ||
| 7 | RBPMS | 1022 | 2.269 | 0.0389 | No | ||
| 8 | AARS | 1142 | 2.196 | 0.0437 | No | ||
| 9 | SNRPB | 1442 | 2.044 | 0.0381 | No | ||
| 10 | NOLA2 | 1834 | 1.891 | 0.0266 | No | ||
| 11 | BICD1 | 2188 | 1.754 | 0.0165 | No | ||
| 12 | UPF1 | 2648 | 1.621 | -0.0000 | No | ||
| 13 | SRRM1 | 2869 | 1.556 | -0.0039 | No | ||
| 14 | NUFIP1 | 3206 | 1.467 | -0.0146 | No | ||
| 15 | GEMIN4 | 3941 | 1.299 | -0.0476 | No | ||
| 16 | PABPC4 | 4396 | 1.200 | -0.0660 | No | ||
| 17 | DDX54 | 4973 | 1.085 | -0.0916 | No | ||
| 18 | RNPS1 | 5670 | 0.953 | -0.1244 | No | ||
| 19 | RRP9 | 5926 | 0.904 | -0.1335 | No | ||
| 20 | PRPF4B | 6208 | 0.852 | -0.1443 | No | ||
| 21 | TXNL4A | 6847 | 0.735 | -0.1751 | No | ||
| 22 | GEMIN6 | 7043 | 0.707 | -0.1820 | No | ||
| 23 | NUDT21 | 7286 | 0.666 | -0.1917 | No | ||
| 24 | SSB | 7844 | 0.559 | -0.2189 | No | ||
| 25 | LSM3 | 7905 | 0.547 | -0.2194 | No | ||
| 26 | NOL3 | 7936 | 0.540 | -0.2182 | No | ||
| 27 | U2AF1 | 8030 | 0.525 | -0.2206 | No | ||
| 28 | ELAVL4 | 8121 | 0.507 | -0.2228 | No | ||
| 29 | SF3B4 | 8215 | 0.489 | -0.2253 | No | ||
| 30 | RBMY1A1 | 8269 | 0.479 | -0.2258 | No | ||
| 31 | DHX16 | 8407 | 0.456 | -0.2308 | No | ||
| 32 | FARS2 | 8491 | 0.436 | -0.2331 | No | ||
| 33 | LSM4 | 8897 | 0.364 | -0.2531 | No | ||
| 34 | GEMIN5 | 9038 | 0.336 | -0.2590 | No | ||
| 35 | SNRP70 | 9162 | 0.307 | -0.2640 | No | ||
| 36 | EXOSC3 | 9570 | 0.218 | -0.2849 | No | ||
| 37 | FBL | 9621 | 0.207 | -0.2866 | No | ||
| 38 | LSM5 | 9635 | 0.202 | -0.2862 | No | ||
| 39 | BCAS2 | 9890 | 0.149 | -0.2992 | No | ||
| 40 | SRPK1 | 10415 | 0.038 | -0.3273 | No | ||
| 41 | RPS14 | 10995 | -0.092 | -0.3582 | No | ||
| 42 | DHX8 | 11069 | -0.110 | -0.3616 | No | ||
| 43 | HNRPF | 11111 | -0.115 | -0.3632 | No | ||
| 44 | SFRS9 | 11352 | -0.172 | -0.3753 | No | ||
| 45 | EXOSC2 | 11533 | -0.215 | -0.3839 | No | ||
| 46 | NONO | 11657 | -0.241 | -0.3893 | No | ||
| 47 | SNRPB2 | 11731 | -0.260 | -0.3920 | No | ||
| 48 | NOLC1 | 12207 | -0.371 | -0.4157 | No | ||
| 49 | PRPF31 | 12341 | -0.405 | -0.4208 | No | ||
| 50 | NOL5A | 12392 | -0.416 | -0.4214 | No | ||
| 51 | PTBP1 | 12809 | -0.523 | -0.4412 | No | ||
| 52 | CPSF3 | 12862 | -0.536 | -0.4413 | No | ||
| 53 | PPAN | 12977 | -0.569 | -0.4445 | No | ||
| 54 | SNRPG | 13132 | -0.612 | -0.4497 | No | ||
| 55 | SIP1 | 13741 | -0.797 | -0.4785 | No | ||
| 56 | SNRPD1 | 13770 | -0.803 | -0.4759 | No | ||
| 57 | EXOSC7 | 14056 | -0.884 | -0.4868 | No | ||
| 58 | PPARGC1A | 14078 | -0.893 | -0.4834 | No | ||
| 59 | RBMS1 | 14194 | -0.931 | -0.4848 | No | ||
| 60 | SNRPF | 14374 | -0.989 | -0.4894 | No | ||
| 61 | DDX39 | 14644 | -1.074 | -0.4985 | No | ||
| 62 | EFTUD2 | 14861 | -1.156 | -0.5042 | No | ||
| 63 | SNRPD3 | 15028 | -1.216 | -0.5070 | Yes | ||
| 64 | SF3A1 | 15078 | -1.237 | -0.5033 | Yes | ||
| 65 | SFRS7 | 15210 | -1.289 | -0.5038 | Yes | ||
| 66 | ZNF346 | 15214 | -1.291 | -0.4973 | Yes | ||
| 67 | CSTF1 | 15269 | -1.315 | -0.4935 | Yes | ||
| 68 | FUSIP1 | 15328 | -1.334 | -0.4898 | Yes | ||
| 69 | SLBP | 15367 | -1.349 | -0.4850 | Yes | ||
| 70 | SNW1 | 15422 | -1.372 | -0.4808 | Yes | ||
| 71 | SF3B2 | 15670 | -1.478 | -0.4866 | Yes | ||
| 72 | SNRPA1 | 15681 | -1.483 | -0.4796 | Yes | ||
| 73 | HNRPD | 15701 | -1.491 | -0.4730 | Yes | ||
| 74 | ADAT1 | 15792 | -1.529 | -0.4700 | Yes | ||
| 75 | NCBP2 | 15840 | -1.552 | -0.4646 | Yes | ||
| 76 | SFRS5 | 15877 | -1.569 | -0.4585 | Yes | ||
| 77 | PRPF8 | 16092 | -1.668 | -0.4615 | Yes | ||
| 78 | SF3B3 | 16119 | -1.680 | -0.4543 | Yes | ||
| 79 | IVNS1ABP | 16132 | -1.685 | -0.4464 | Yes | ||
| 80 | ASCC3L1 | 16160 | -1.696 | -0.4391 | Yes | ||
| 81 | DGCR8 | 16322 | -1.773 | -0.4387 | Yes | ||
| 82 | PABPC1 | 16324 | -1.775 | -0.4297 | Yes | ||
| 83 | CPSF1 | 16348 | -1.786 | -0.4218 | Yes | ||
| 84 | RBM5 | 16365 | -1.792 | -0.4135 | Yes | ||
| 85 | HNRPUL1 | 16657 | -1.935 | -0.4193 | Yes | ||
| 86 | SFRS10 | 16701 | -1.959 | -0.4116 | Yes | ||
| 87 | KHDRBS1 | 16794 | -2.018 | -0.4062 | Yes | ||
| 88 | NSUN2 | 16906 | -2.069 | -0.4016 | Yes | ||
| 89 | TRNT1 | 16992 | -2.116 | -0.3954 | Yes | ||
| 90 | RNMT | 17000 | -2.120 | -0.3849 | Yes | ||
| 91 | SPOP | 17008 | -2.123 | -0.3744 | Yes | ||
| 92 | HNRPDL | 17024 | -2.135 | -0.3643 | Yes | ||
| 93 | PHF5A | 17060 | -2.150 | -0.3552 | Yes | ||
| 94 | USP39 | 17247 | -2.259 | -0.3537 | Yes | ||
| 95 | SF3A3 | 17334 | -2.311 | -0.3465 | Yes | ||
| 96 | RBM3 | 17351 | -2.322 | -0.3354 | Yes | ||
| 97 | INTS6 | 17365 | -2.327 | -0.3242 | Yes | ||
| 98 | SYNCRIP | 17454 | -2.383 | -0.3168 | Yes | ||
| 99 | SFRS2 | 17548 | -2.439 | -0.3093 | Yes | ||
| 100 | POP4 | 17635 | -2.494 | -0.3012 | Yes | ||
| 101 | RBM6 | 17679 | -2.526 | -0.2906 | Yes | ||
| 102 | DHX38 | 17691 | -2.532 | -0.2782 | Yes | ||
| 103 | RNGTT | 17757 | -2.575 | -0.2685 | Yes | ||
| 104 | GRSF1 | 17827 | -2.620 | -0.2588 | Yes | ||
| 105 | CUGBP1 | 17880 | -2.670 | -0.2480 | Yes | ||
| 106 | SFRS2IP | 17936 | -2.710 | -0.2371 | Yes | ||
| 107 | DHX15 | 18058 | -2.822 | -0.2291 | Yes | ||
| 108 | ZNF638 | 18075 | -2.833 | -0.2155 | Yes | ||
| 109 | CSTF3 | 18086 | -2.844 | -0.2015 | Yes | ||
| 110 | CDC2L2 | 18111 | -2.871 | -0.1881 | Yes | ||
| 111 | SNRPN | 18195 | -2.969 | -0.1773 | Yes | ||
| 112 | SFPQ | 18263 | -3.078 | -0.1652 | Yes | ||
| 113 | SRPK2 | 18286 | -3.115 | -0.1504 | Yes | ||
| 114 | DUSP11 | 18288 | -3.117 | -0.1345 | Yes | ||
| 115 | SFRS1 | 18333 | -3.183 | -0.1206 | Yes | ||
| 116 | CSTF2 | 18338 | -3.191 | -0.1045 | Yes | ||
| 117 | SFRS6 | 18350 | -3.206 | -0.0886 | Yes | ||
| 118 | CRNKL1 | 18394 | -3.286 | -0.0741 | Yes | ||
| 119 | KIN | 18482 | -3.536 | -0.0607 | Yes | ||
| 120 | DDX20 | 18519 | -3.704 | -0.0437 | Yes | ||
| 121 | DBR1 | 18583 | -4.235 | -0.0254 | Yes | ||
| 122 | PRPF3 | 18608 | -5.298 | 0.0004 | Yes |
