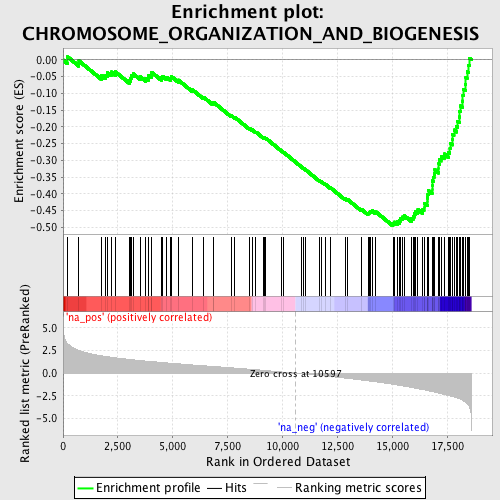
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHROMOSOME_ORGANIZATION_AND_BIOGENESIS |
| Enrichment Score (ES) | -0.4962437 |
| Normalized Enrichment Score (NES) | -2.2898383 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0013857095 |
| FWER p-Value | 0.0050 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HTATIP | 206 | 3.244 | 0.0093 | No | ||
| 2 | ERCC1 | 721 | 2.491 | -0.0028 | No | ||
| 3 | HMGA1 | 1742 | 1.926 | -0.0458 | No | ||
| 4 | AIFM2 | 1945 | 1.845 | -0.0451 | No | ||
| 5 | SMARCD1 | 2029 | 1.813 | -0.0382 | No | ||
| 6 | MTA2 | 2190 | 1.753 | -0.0358 | No | ||
| 7 | RBM14 | 2381 | 1.692 | -0.0354 | No | ||
| 8 | HDAC6 | 3020 | 1.516 | -0.0603 | No | ||
| 9 | CHMP1A | 3080 | 1.502 | -0.0541 | No | ||
| 10 | CENPE | 3121 | 1.491 | -0.0469 | No | ||
| 11 | RBBP4 | 3192 | 1.470 | -0.0414 | No | ||
| 12 | HDAC5 | 3516 | 1.394 | -0.0501 | No | ||
| 13 | HDAC3 | 3770 | 1.334 | -0.0554 | No | ||
| 14 | HDAC7A | 3879 | 1.310 | -0.0529 | No | ||
| 15 | MYST3 | 3903 | 1.304 | -0.0460 | No | ||
| 16 | TSSK6 | 4026 | 1.282 | -0.0445 | No | ||
| 17 | SIRT2 | 4034 | 1.282 | -0.0368 | No | ||
| 18 | NAP1L3 | 4473 | 1.183 | -0.0530 | No | ||
| 19 | SUPT4H1 | 4536 | 1.168 | -0.0491 | No | ||
| 20 | BNIP3 | 4730 | 1.129 | -0.0524 | No | ||
| 21 | MYST1 | 4901 | 1.097 | -0.0547 | No | ||
| 22 | GCN5L2 | 4942 | 1.090 | -0.0500 | No | ||
| 23 | TEP1 | 5247 | 1.032 | -0.0599 | No | ||
| 24 | TINF2 | 5897 | 0.911 | -0.0892 | No | ||
| 25 | ACIN1 | 6405 | 0.819 | -0.1114 | No | ||
| 26 | JMJD2A | 6831 | 0.737 | -0.1298 | No | ||
| 27 | SYCP3 | 6863 | 0.732 | -0.1268 | No | ||
| 28 | TAF6L | 7652 | 0.596 | -0.1656 | No | ||
| 29 | SET | 7829 | 0.561 | -0.1716 | No | ||
| 30 | PHB | 8480 | 0.439 | -0.2040 | No | ||
| 31 | TERT | 8610 | 0.414 | -0.2083 | No | ||
| 32 | STAG3 | 8789 | 0.383 | -0.2155 | No | ||
| 33 | SIRT5 | 9149 | 0.311 | -0.2330 | No | ||
| 34 | MSH2 | 9177 | 0.304 | -0.2325 | No | ||
| 35 | CHAF1B | 9230 | 0.291 | -0.2335 | No | ||
| 36 | HDAC10 | 9965 | 0.134 | -0.2723 | No | ||
| 37 | DFFB | 10059 | 0.117 | -0.2766 | No | ||
| 38 | CHAF1A | 10863 | -0.064 | -0.3195 | No | ||
| 39 | CARM1 | 10967 | -0.087 | -0.3246 | No | ||
| 40 | CDCA5 | 11033 | -0.101 | -0.3274 | No | ||
| 41 | DDX11 | 11673 | -0.244 | -0.3604 | No | ||
| 42 | POT1 | 11773 | -0.269 | -0.3641 | No | ||
| 43 | PAM | 11949 | -0.310 | -0.3716 | No | ||
| 44 | HDAC8 | 12185 | -0.367 | -0.3819 | No | ||
| 45 | NAP1L2 | 12852 | -0.534 | -0.4146 | No | ||
| 46 | HDAC11 | 12960 | -0.562 | -0.4168 | No | ||
| 47 | SUV39H2 | 13591 | -0.749 | -0.4461 | No | ||
| 48 | CDC23 | 13900 | -0.834 | -0.4575 | No | ||
| 49 | TNP1 | 13948 | -0.848 | -0.4547 | No | ||
| 50 | MAP3K12 | 13991 | -0.861 | -0.4516 | No | ||
| 51 | PPARGC1A | 14078 | -0.893 | -0.4506 | No | ||
| 52 | NPM2 | 14257 | -0.950 | -0.4542 | No | ||
| 53 | HMGA2 | 15036 | -1.219 | -0.4886 | Yes | ||
| 54 | SMG6 | 15104 | -1.250 | -0.4843 | Yes | ||
| 55 | BPTF | 15228 | -1.299 | -0.4828 | Yes | ||
| 56 | CREBBP | 15329 | -1.334 | -0.4798 | Yes | ||
| 57 | HELLS | 15364 | -1.348 | -0.4732 | Yes | ||
| 58 | PTGES3 | 15456 | -1.389 | -0.4694 | Yes | ||
| 59 | SUPT16H | 15557 | -1.427 | -0.4658 | Yes | ||
| 60 | HDAC1 | 15872 | -1.567 | -0.4729 | Yes | ||
| 61 | MSH3 | 15952 | -1.600 | -0.4671 | Yes | ||
| 62 | SMC4 | 15999 | -1.626 | -0.4594 | Yes | ||
| 63 | MRE11A | 16048 | -1.648 | -0.4516 | Yes | ||
| 64 | ZWINT | 16172 | -1.700 | -0.4475 | Yes | ||
| 65 | ASF1A | 16393 | -1.808 | -0.4481 | Yes | ||
| 66 | EZH2 | 16450 | -1.831 | -0.4396 | Yes | ||
| 67 | ACTL6A | 16453 | -1.832 | -0.4282 | Yes | ||
| 68 | SMARCC1 | 16602 | -1.911 | -0.4241 | Yes | ||
| 69 | NAP1L4 | 16604 | -1.914 | -0.4121 | Yes | ||
| 70 | MYST4 | 16623 | -1.922 | -0.4010 | Yes | ||
| 71 | TOP2A | 16650 | -1.933 | -0.3903 | Yes | ||
| 72 | ZW10 | 16818 | -2.028 | -0.3865 | Yes | ||
| 73 | SMARCE1 | 16820 | -2.028 | -0.3738 | Yes | ||
| 74 | NAP1L1 | 16827 | -2.032 | -0.3614 | Yes | ||
| 75 | SIRT1 | 16877 | -2.056 | -0.3511 | Yes | ||
| 76 | NSD1 | 16908 | -2.069 | -0.3397 | Yes | ||
| 77 | RPS6KA5 | 16943 | -2.089 | -0.3284 | Yes | ||
| 78 | UBE2N | 17123 | -2.188 | -0.3243 | Yes | ||
| 79 | ERCC4 | 17126 | -2.190 | -0.3107 | Yes | ||
| 80 | ARID1A | 17156 | -2.208 | -0.2983 | Yes | ||
| 81 | NBN | 17226 | -2.246 | -0.2879 | Yes | ||
| 82 | CENPH | 17368 | -2.331 | -0.2809 | Yes | ||
| 83 | SYCP1 | 17560 | -2.444 | -0.2758 | Yes | ||
| 84 | RFC1 | 17631 | -2.492 | -0.2640 | Yes | ||
| 85 | HMGB1 | 17643 | -2.502 | -0.2488 | Yes | ||
| 86 | NASP | 17744 | -2.569 | -0.2381 | Yes | ||
| 87 | TLK1 | 17754 | -2.574 | -0.2224 | Yes | ||
| 88 | TLK2 | 17816 | -2.616 | -0.2092 | Yes | ||
| 89 | HDAC2 | 17934 | -2.708 | -0.1985 | Yes | ||
| 90 | NUSAP1 | 17980 | -2.749 | -0.1836 | Yes | ||
| 91 | POLS | 18062 | -2.824 | -0.1703 | Yes | ||
| 92 | RSF1 | 18074 | -2.833 | -0.1530 | Yes | ||
| 93 | SMARCA5 | 18120 | -2.882 | -0.1373 | Yes | ||
| 94 | RAD50 | 18194 | -2.966 | -0.1226 | Yes | ||
| 95 | TERF2 | 18220 | -3.011 | -0.1051 | Yes | ||
| 96 | TERF2IP | 18253 | -3.068 | -0.0875 | Yes | ||
| 97 | SMC1A | 18339 | -3.191 | -0.0720 | Yes | ||
| 98 | NCAPH | 18356 | -3.228 | -0.0526 | Yes | ||
| 99 | SATB1 | 18414 | -3.343 | -0.0346 | Yes | ||
| 100 | NDC80 | 18478 | -3.521 | -0.0159 | Yes | ||
| 101 | EHMT1 | 18518 | -3.704 | 0.0053 | Yes |