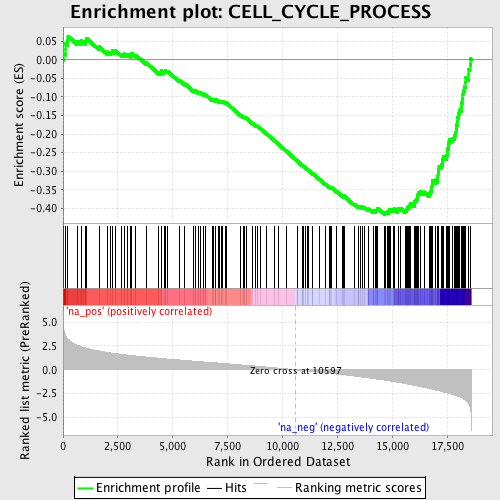
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CELL_CYCLE_PROCESS |
| Enrichment Score (ES) | -0.41660008 |
| Normalized Enrichment Score (NES) | -2.0583959 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00824881 |
| FWER p-Value | 0.103 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CDK10 | 30 | 4.309 | 0.0164 | No | ||
| 2 | BOLL | 99 | 3.635 | 0.0279 | No | ||
| 3 | CDK4 | 103 | 3.623 | 0.0429 | No | ||
| 4 | ABL1 | 205 | 3.245 | 0.0511 | No | ||
| 5 | BCAT1 | 221 | 3.210 | 0.0637 | No | ||
| 6 | ANAPC11 | 662 | 2.548 | 0.0505 | No | ||
| 7 | RAD51L3 | 820 | 2.422 | 0.0521 | No | ||
| 8 | TBRG4 | 1026 | 2.266 | 0.0505 | No | ||
| 9 | DCTN2 | 1071 | 2.241 | 0.0575 | No | ||
| 10 | PML | 1644 | 1.966 | 0.0348 | No | ||
| 11 | CDKN2C | 2038 | 1.811 | 0.0210 | No | ||
| 12 | MSH5 | 2180 | 1.758 | 0.0208 | No | ||
| 13 | SSSCA1 | 2252 | 1.731 | 0.0242 | No | ||
| 14 | MAD2L2 | 2384 | 1.691 | 0.0242 | No | ||
| 15 | NEK2 | 2671 | 1.615 | 0.0154 | No | ||
| 16 | CDC25B | 2787 | 1.580 | 0.0158 | No | ||
| 17 | ACVR1B | 2923 | 1.541 | 0.0149 | No | ||
| 18 | CHMP1A | 3080 | 1.502 | 0.0128 | No | ||
| 19 | CENPE | 3121 | 1.491 | 0.0168 | No | ||
| 20 | PLK1 | 3309 | 1.442 | 0.0128 | No | ||
| 21 | CETN1 | 3783 | 1.331 | -0.0073 | No | ||
| 22 | MAP3K11 | 4368 | 1.207 | -0.0339 | No | ||
| 23 | CDKN1B | 4465 | 1.185 | -0.0341 | No | ||
| 24 | APBB1 | 4471 | 1.184 | -0.0294 | No | ||
| 25 | DMC1 | 4599 | 1.155 | -0.0315 | No | ||
| 26 | CUL2 | 4647 | 1.147 | -0.0292 | No | ||
| 27 | TPD52L1 | 4758 | 1.124 | -0.0305 | No | ||
| 28 | TTN | 5301 | 1.022 | -0.0556 | No | ||
| 29 | CUL5 | 5535 | 0.976 | -0.0641 | No | ||
| 30 | NEK6 | 5956 | 0.899 | -0.0831 | No | ||
| 31 | CDK6 | 6023 | 0.888 | -0.0830 | No | ||
| 32 | TIMELESS | 6161 | 0.862 | -0.0868 | No | ||
| 33 | POLD1 | 6261 | 0.843 | -0.0886 | No | ||
| 34 | PIM2 | 6386 | 0.822 | -0.0919 | No | ||
| 35 | KATNA1 | 6467 | 0.807 | -0.0929 | No | ||
| 36 | DUSP13 | 6787 | 0.747 | -0.1070 | No | ||
| 37 | CDKN2B | 6856 | 0.733 | -0.1076 | No | ||
| 38 | CENTD2 | 6941 | 0.722 | -0.1092 | No | ||
| 39 | CDKN3 | 6964 | 0.719 | -0.1073 | No | ||
| 40 | NUMA1 | 7085 | 0.700 | -0.1109 | No | ||
| 41 | KIF11 | 7145 | 0.690 | -0.1112 | No | ||
| 42 | CD28 | 7225 | 0.675 | -0.1127 | No | ||
| 43 | APBB2 | 7274 | 0.667 | -0.1125 | No | ||
| 44 | ACVR1 | 7384 | 0.648 | -0.1157 | No | ||
| 45 | TOP3A | 7424 | 0.640 | -0.1151 | No | ||
| 46 | NPM1 | 8095 | 0.513 | -0.1492 | No | ||
| 47 | CLIP1 | 8232 | 0.487 | -0.1546 | No | ||
| 48 | MSH4 | 8274 | 0.479 | -0.1548 | No | ||
| 49 | TGFB1 | 8358 | 0.463 | -0.1574 | No | ||
| 50 | RAD1 | 8641 | 0.409 | -0.1709 | No | ||
| 51 | STAG3 | 8789 | 0.383 | -0.1773 | No | ||
| 52 | POLA1 | 8844 | 0.374 | -0.1786 | No | ||
| 53 | PRMT5 | 8974 | 0.349 | -0.1842 | No | ||
| 54 | E2F1 | 9255 | 0.288 | -0.1981 | No | ||
| 55 | RCC1 | 9622 | 0.206 | -0.2171 | No | ||
| 56 | CDKN2D | 9831 | 0.160 | -0.2277 | No | ||
| 57 | BUB1 | 10189 | 0.087 | -0.2467 | No | ||
| 58 | UBE2C | 10662 | -0.017 | -0.2722 | No | ||
| 59 | CCNA1 | 10686 | -0.025 | -0.2733 | No | ||
| 60 | EREG | 10902 | -0.073 | -0.2847 | No | ||
| 61 | RAD52 | 10916 | -0.076 | -0.2850 | No | ||
| 62 | RAD54L | 10953 | -0.084 | -0.2866 | No | ||
| 63 | CDCA5 | 11033 | -0.101 | -0.2905 | No | ||
| 64 | CENPF | 11128 | -0.120 | -0.2951 | No | ||
| 65 | KIF2C | 11196 | -0.138 | -0.2981 | No | ||
| 66 | DCTN3 | 11376 | -0.178 | -0.3071 | No | ||
| 67 | MDM4 | 11377 | -0.178 | -0.3064 | No | ||
| 68 | DDX11 | 11673 | -0.244 | -0.3213 | No | ||
| 69 | PAM | 11949 | -0.310 | -0.3349 | No | ||
| 70 | GFI1 | 12138 | -0.356 | -0.3436 | No | ||
| 71 | TUBG1 | 12162 | -0.363 | -0.3433 | No | ||
| 72 | NOLC1 | 12207 | -0.371 | -0.3442 | No | ||
| 73 | KIF22 | 12230 | -0.378 | -0.3438 | No | ||
| 74 | TUBE1 | 12482 | -0.443 | -0.3555 | No | ||
| 75 | TARDBP | 12728 | -0.503 | -0.3667 | No | ||
| 76 | CHEK1 | 12792 | -0.517 | -0.3679 | No | ||
| 77 | CDKN1A | 12803 | -0.522 | -0.3663 | No | ||
| 78 | PPP5C | 13288 | -0.656 | -0.3898 | No | ||
| 79 | XRCC2 | 13445 | -0.703 | -0.3953 | No | ||
| 80 | RAN | 13479 | -0.712 | -0.3941 | No | ||
| 81 | BRCA2 | 13576 | -0.743 | -0.3962 | No | ||
| 82 | DLG7 | 13634 | -0.762 | -0.3961 | No | ||
| 83 | STMN1 | 13732 | -0.793 | -0.3980 | No | ||
| 84 | CDC23 | 13900 | -0.834 | -0.4036 | No | ||
| 85 | CDC6 | 13931 | -0.843 | -0.4017 | No | ||
| 86 | ATM | 14127 | -0.911 | -0.4084 | No | ||
| 87 | BIRC5 | 14134 | -0.913 | -0.4049 | No | ||
| 88 | NPM2 | 14257 | -0.950 | -0.4076 | No | ||
| 89 | TTK | 14282 | -0.959 | -0.4048 | No | ||
| 90 | ANAPC10 | 14310 | -0.967 | -0.4023 | No | ||
| 91 | EPGN | 14336 | -0.975 | -0.3995 | No | ||
| 92 | GFI1B | 14652 | -1.077 | -0.4121 | Yes | ||
| 93 | CDK2AP1 | 14701 | -1.100 | -0.4101 | Yes | ||
| 94 | CDC16 | 14779 | -1.128 | -0.4095 | Yes | ||
| 95 | KNTC1 | 14846 | -1.150 | -0.4083 | Yes | ||
| 96 | CIT | 14854 | -1.153 | -0.4039 | Yes | ||
| 97 | PCBP4 | 14926 | -1.177 | -0.4028 | Yes | ||
| 98 | AURKA | 15058 | -1.229 | -0.4047 | Yes | ||
| 99 | TIPIN | 15091 | -1.244 | -0.4013 | Yes | ||
| 100 | SUGT1 | 15263 | -1.313 | -0.4050 | Yes | ||
| 101 | HSPA2 | 15297 | -1.325 | -0.4013 | Yes | ||
| 102 | RACGAP1 | 15365 | -1.348 | -0.3993 | Yes | ||
| 103 | USH1C | 15583 | -1.439 | -0.4050 | Yes | ||
| 104 | PPP6C | 15643 | -1.463 | -0.4021 | Yes | ||
| 105 | MYH10 | 15683 | -1.483 | -0.3980 | Yes | ||
| 106 | LIG3 | 15734 | -1.505 | -0.3944 | Yes | ||
| 107 | ANAPC5 | 15804 | -1.535 | -0.3917 | Yes | ||
| 108 | NDE1 | 15842 | -1.552 | -0.3872 | Yes | ||
| 109 | SMC4 | 15999 | -1.626 | -0.3888 | Yes | ||
| 110 | GSPT1 | 16003 | -1.629 | -0.3822 | Yes | ||
| 111 | MRE11A | 16048 | -1.648 | -0.3777 | Yes | ||
| 112 | PIN1 | 16127 | -1.683 | -0.3748 | Yes | ||
| 113 | CDKN1C | 16139 | -1.686 | -0.3684 | Yes | ||
| 114 | ZWINT | 16172 | -1.700 | -0.3630 | Yes | ||
| 115 | FBXO5 | 16196 | -1.714 | -0.3571 | Yes | ||
| 116 | CDC25C | 16298 | -1.762 | -0.3552 | Yes | ||
| 117 | MPHOSPH6 | 16456 | -1.833 | -0.3560 | Yes | ||
| 118 | CETN3 | 16690 | -1.953 | -0.3604 | Yes | ||
| 119 | RAD51 | 16726 | -1.978 | -0.3541 | Yes | ||
| 120 | KHDRBS1 | 16794 | -2.018 | -0.3492 | Yes | ||
| 121 | CDKN2A | 16805 | -2.022 | -0.3413 | Yes | ||
| 122 | ZW10 | 16818 | -2.028 | -0.3335 | Yes | ||
| 123 | CUL1 | 16836 | -2.035 | -0.3259 | Yes | ||
| 124 | CHFR | 16958 | -2.095 | -0.3237 | Yes | ||
| 125 | RAD17 | 17065 | -2.152 | -0.3204 | Yes | ||
| 126 | PAFAH1B1 | 17084 | -2.165 | -0.3123 | Yes | ||
| 127 | RAD21 | 17089 | -2.167 | -0.3035 | Yes | ||
| 128 | SKP2 | 17105 | -2.174 | -0.2952 | Yes | ||
| 129 | PTPRC | 17131 | -2.196 | -0.2873 | Yes | ||
| 130 | NBN | 17226 | -2.246 | -0.2830 | Yes | ||
| 131 | SMC3 | 17288 | -2.281 | -0.2768 | Yes | ||
| 132 | RAD51L1 | 17300 | -2.290 | -0.2678 | Yes | ||
| 133 | RB1 | 17323 | -2.301 | -0.2594 | Yes | ||
| 134 | CDK2 | 17488 | -2.401 | -0.2582 | Yes | ||
| 135 | KIF23 | 17511 | -2.417 | -0.2493 | Yes | ||
| 136 | ANAPC4 | 17523 | -2.423 | -0.2397 | Yes | ||
| 137 | SYCP1 | 17560 | -2.444 | -0.2314 | Yes | ||
| 138 | POLE | 17578 | -2.455 | -0.2221 | Yes | ||
| 139 | PRC1 | 17622 | -2.488 | -0.2140 | Yes | ||
| 140 | KPNA2 | 17761 | -2.577 | -0.2107 | Yes | ||
| 141 | BUB1B | 17842 | -2.637 | -0.2040 | Yes | ||
| 142 | MAD2L1 | 17898 | -2.685 | -0.1957 | Yes | ||
| 143 | CUL3 | 17909 | -2.691 | -0.1850 | Yes | ||
| 144 | AKAP8 | 17942 | -2.714 | -0.1754 | Yes | ||
| 145 | SPO11 | 17961 | -2.732 | -0.1649 | Yes | ||
| 146 | NUSAP1 | 17980 | -2.749 | -0.1544 | Yes | ||
| 147 | EGF | 18031 | -2.793 | -0.1454 | Yes | ||
| 148 | POLS | 18062 | -2.824 | -0.1352 | Yes | ||
| 149 | TPX2 | 18142 | -2.908 | -0.1273 | Yes | ||
| 150 | CDC7 | 18162 | -2.928 | -0.1161 | Yes | ||
| 151 | RAD50 | 18194 | -2.966 | -0.1053 | Yes | ||
| 152 | KIF15 | 18215 | -2.995 | -0.0939 | Yes | ||
| 153 | CCNA2 | 18230 | -3.023 | -0.0820 | Yes | ||
| 154 | CUL4A | 18284 | -3.111 | -0.0718 | Yes | ||
| 155 | SMC1A | 18339 | -3.191 | -0.0614 | Yes | ||
| 156 | NCAPH | 18356 | -3.228 | -0.0488 | Yes | ||
| 157 | LATS2 | 18471 | -3.501 | -0.0403 | Yes | ||
| 158 | NDC80 | 18478 | -3.521 | -0.0259 | Yes | ||
| 159 | ANLN | 18546 | -3.873 | -0.0133 | Yes | ||
| 160 | TGFA | 18567 | -4.065 | 0.0027 | Yes |
