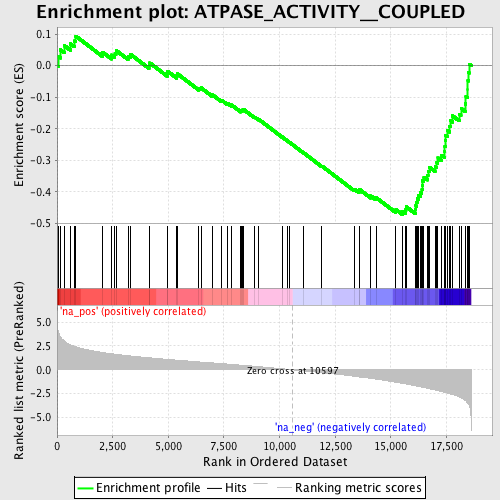
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATPASE_ACTIVITY__COUPLED |
| Enrichment Score (ES) | -0.47202104 |
| Normalized Enrichment Score (NES) | -2.0930858 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008222469 |
| FWER p-Value | 0.071 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 48 | 4.113 | 0.0292 | No | ||
| 2 | ERCC8 | 142 | 3.406 | 0.0505 | No | ||
| 3 | PEX6 | 321 | 2.993 | 0.0640 | No | ||
| 4 | SKIV2L | 598 | 2.619 | 0.0693 | No | ||
| 5 | MYL6 | 780 | 2.450 | 0.0785 | No | ||
| 6 | TAPBP | 841 | 2.399 | 0.0937 | No | ||
| 7 | ATP1A3 | 2049 | 1.807 | 0.0426 | No | ||
| 8 | RUVBL2 | 2450 | 1.673 | 0.0339 | No | ||
| 9 | ATP2A1 | 2601 | 1.635 | 0.0385 | No | ||
| 10 | UPF1 | 2648 | 1.621 | 0.0485 | No | ||
| 11 | RBBP4 | 3192 | 1.470 | 0.0306 | No | ||
| 12 | ABCC3 | 3305 | 1.443 | 0.0357 | No | ||
| 13 | MYH6 | 4133 | 1.260 | 0.0008 | No | ||
| 14 | ATP1A2 | 4168 | 1.252 | 0.0086 | No | ||
| 15 | ABCG1 | 4946 | 1.089 | -0.0249 | No | ||
| 16 | DDX54 | 4973 | 1.085 | -0.0179 | No | ||
| 17 | ABCD4 | 5379 | 1.007 | -0.0320 | No | ||
| 18 | ATP2B3 | 5398 | 1.002 | -0.0252 | No | ||
| 19 | ABCB11 | 6372 | 0.826 | -0.0713 | No | ||
| 20 | KATNA1 | 6467 | 0.807 | -0.0701 | No | ||
| 21 | ATP4B | 6986 | 0.716 | -0.0925 | No | ||
| 22 | ABCC2 | 7388 | 0.648 | -0.1092 | No | ||
| 23 | ABCC1 | 7651 | 0.596 | -0.1187 | No | ||
| 24 | ATP6V0E1 | 7838 | 0.560 | -0.1244 | No | ||
| 25 | MYO3A | 8259 | 0.481 | -0.1433 | No | ||
| 26 | XRCC5 | 8298 | 0.473 | -0.1417 | No | ||
| 27 | MYO1E | 8342 | 0.466 | -0.1405 | No | ||
| 28 | ATP6V0C | 8380 | 0.460 | -0.1389 | No | ||
| 29 | EIF4A3 | 8880 | 0.368 | -0.1630 | No | ||
| 30 | ABCG2 | 9058 | 0.330 | -0.1700 | No | ||
| 31 | ABCA3 | 10110 | 0.106 | -0.2258 | No | ||
| 32 | SMARCA1 | 10344 | 0.053 | -0.2380 | No | ||
| 33 | RFC3 | 10437 | 0.034 | -0.2427 | No | ||
| 34 | DHX8 | 11069 | -0.110 | -0.2759 | No | ||
| 35 | ATP7A | 11898 | -0.299 | -0.3182 | No | ||
| 36 | DDX56 | 13360 | -0.679 | -0.3918 | No | ||
| 37 | MYH7 | 13574 | -0.741 | -0.3976 | No | ||
| 38 | CHD2 | 13584 | -0.746 | -0.3923 | No | ||
| 39 | ATP2C1 | 14098 | -0.902 | -0.4130 | No | ||
| 40 | CCT8 | 14337 | -0.976 | -0.4183 | No | ||
| 41 | BPTF | 15228 | -1.299 | -0.4563 | No | ||
| 42 | DHX9 | 15521 | -1.413 | -0.4611 | Yes | ||
| 43 | FXYD2 | 15663 | -1.475 | -0.4573 | Yes | ||
| 44 | MYH10 | 15683 | -1.483 | -0.4469 | Yes | ||
| 45 | ABCC6 | 16110 | -1.678 | -0.4569 | Yes | ||
| 46 | DDX3X | 16124 | -1.681 | -0.4446 | Yes | ||
| 47 | ASCC3L1 | 16160 | -1.696 | -0.4334 | Yes | ||
| 48 | ATP1B2 | 16186 | -1.708 | -0.4216 | Yes | ||
| 49 | ABCF1 | 16250 | -1.733 | -0.4116 | Yes | ||
| 50 | ATP1A1 | 16336 | -1.782 | -0.4024 | Yes | ||
| 51 | ATP1B1 | 16399 | -1.812 | -0.3918 | Yes | ||
| 52 | VPS4B | 16416 | -1.820 | -0.3786 | Yes | ||
| 53 | SMARCAL1 | 16430 | -1.823 | -0.3652 | Yes | ||
| 54 | ATP4A | 16488 | -1.849 | -0.3540 | Yes | ||
| 55 | TOP2A | 16650 | -1.933 | -0.3478 | Yes | ||
| 56 | ATP6V1B2 | 16676 | -1.944 | -0.3341 | Yes | ||
| 57 | RAD51 | 16726 | -1.978 | -0.3215 | Yes | ||
| 58 | CHD4 | 16990 | -2.116 | -0.3193 | Yes | ||
| 59 | DDX25 | 17068 | -2.153 | -0.3069 | Yes | ||
| 60 | ABCD3 | 17118 | -2.185 | -0.2926 | Yes | ||
| 61 | MYH9 | 17279 | -2.276 | -0.2837 | Yes | ||
| 62 | DDX21 | 17421 | -2.360 | -0.2731 | Yes | ||
| 63 | ABCB7 | 17430 | -2.367 | -0.2552 | Yes | ||
| 64 | RALBP1 | 17451 | -2.381 | -0.2379 | Yes | ||
| 65 | ATP6V1C1 | 17467 | -2.388 | -0.2203 | Yes | ||
| 66 | ATP1B3 | 17538 | -2.435 | -0.2053 | Yes | ||
| 67 | XRCC6 | 17640 | -2.499 | -0.1914 | Yes | ||
| 68 | DHX38 | 17691 | -2.532 | -0.1746 | Yes | ||
| 69 | G3BP1 | 17775 | -2.585 | -0.1591 | Yes | ||
| 70 | CLPX | 18077 | -2.834 | -0.1534 | Yes | ||
| 71 | ATP7B | 18156 | -2.924 | -0.1351 | Yes | ||
| 72 | DDX18 | 18344 | -3.197 | -0.1205 | Yes | ||
| 73 | RECQL | 18376 | -3.254 | -0.0970 | Yes | ||
| 74 | CHD1 | 18463 | -3.482 | -0.0748 | Yes | ||
| 75 | ATP2A2 | 18465 | -3.491 | -0.0479 | Yes | ||
| 76 | ATP11B | 18483 | -3.537 | -0.0215 | Yes | ||
| 77 | DDX20 | 18519 | -3.704 | 0.0052 | Yes |
